Measurement of chemical relationship by PMD based reactomics
Published in Chemistry
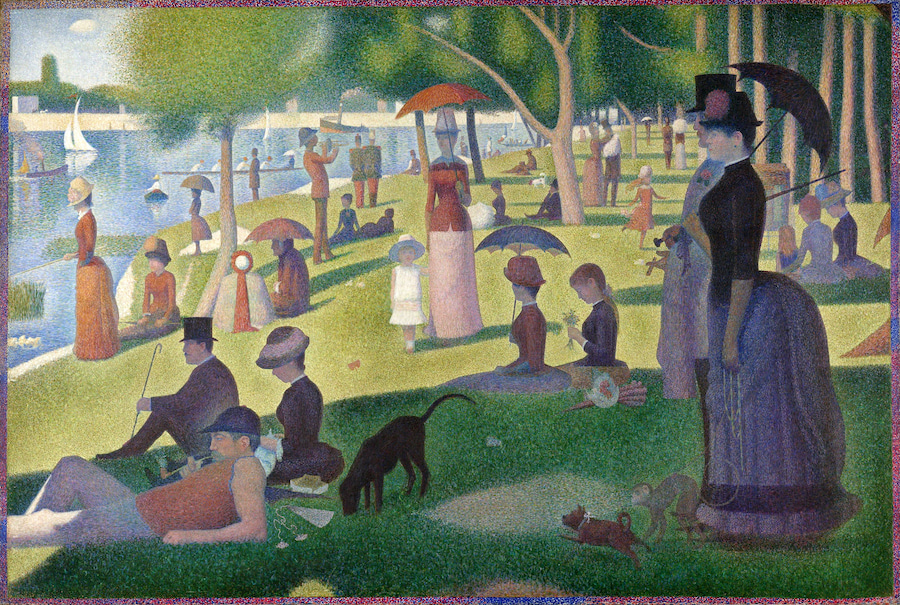
Modern High Resolution Mass Spectrometry (HRMS) has enabled the untargeted detection of tens of thousands of peaks/features from a single sample in a single assay. However, 'untargeted' means that many of these features are of unknown chemical identity. After a chemist isolates and identifies these compounds, a biologist can then build the relationships and interpret the findings based on the experimental design. The workflow is:
peaks/features -> identified compounds -> relationship among compounds
Identification of compounds not only plays an important role to connect the chemical assay to the biological interpretation, but is also a time consuming step, particularly to pursue standards validation. We need to rely on databases of standards and deal with the common situation in which there is no commercially available chemical standard. However, is the identification of compounds required in order to study the relationship among compounds?
Let's have a rest and visit the museum.
This is A Sunday Afternoon on the Island of La Grande Jatte by Georges Seurat (via Wikimedia Commons). Here, we see couples, friends, families ... Wait a minute, but how do you know those relationships without knowing who they are? The answer is "distance". People can be grouped by paired distances, and family members and close friends always sit close together.
Similar things happen in HRMS based untargeted analysis. When we collect peaks from samples, we actually capture a snapshot of all compounds with their paired chemical reaction relationships like the painting. For example, when we see two compounds that show a paired mass distance (PMD) of 15.995Da, those two compounds might be involved in a xenobiotics Phase I reaction such as hydrolysis, reduction and oxidation.
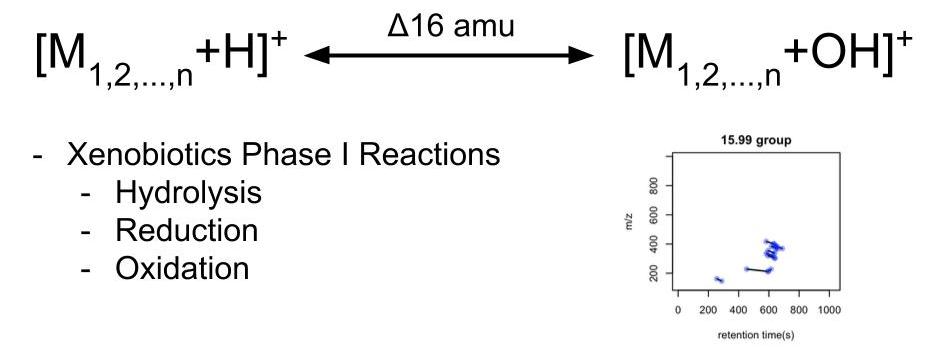
If a lot of paired compounds show this relationship, we could use the sum of their intensities as a proxy for the level of certain reactions in the system. In this case, we could skip the annotation and directly make quantitative analysis at the reactions level.
peaks/features -> paired mass distance -> relationship among compounds.
In the published paper in Communication Chemistry, we analyzed all of the reactions in the KEGG reaction database as well as all the PMDs of the compounds in HMDB. We found that most of the reactions, or PMDs of known reaction/compounds are highly enriched in only a few PMDs. In this case, PMD could serve as a dimension reduction technique to directly check reaction level changes among samples with clear chemical meanings. We call it "PMD based reactomics". We provide a formal definition of qualitative and quantitative analysis to measure reaction level changes by PMD. Three applications of PMD based reactomics were shown: recursive metabolites discovery by PMD network analysis, source appointment of unknown endogenous/exogenous compounds, and biomarker reaction discovery in an example of cancer research. In this latter case, for an untargeted HRMS dataset, we used PMD to reveal reaction level changes in the samples as a new dimension to interpret the data.
We appreciate the open science concept and have released all of the data and code to reproduce our findings. If you are interested in PMD based reactomics, you can also check out our online slides and oral presentation video on ASMS 2020 reboot. We also developed a free and open source R package with a tutorial. Finally, you can also find the open review with our reply when the draft is released on BioRxiv as preprint. We invite you to try PMD analysis and are happy to discuss the next collaboration.
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
f-block chemistry
Publishing Model: Open Access
Deadline: Feb 28, 2026
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in