New insights into the emergence of life’s genetic polymers: The role of hybrid sequences
Published in Chemistry

DNA, RNA, and proteins are central to the current life on Earth where they play the genotypic, phenotypic and catalytic role respectively. But how did these biopolymers arise before biology? The RNA world hypothesis postulates RNA to be the first genetic polymer and protein and DNA are its later descendants. Hence, most of the efforts have been guided by the desire of a clean prebiotic synthesis of RNA (or pre-RNA molecule, which later converted to RNA). However, various studies indicate that other nucleic acids, such as DNA and other pre-RNA molecules (like TNA) could have been available simultaneously on early Earth. Therefore, besides forming their respective homogeneous polymers, the other nucleic acids could also become part of the other polymers and make hybrid (chimeric) sequences as well. Such kind of chimeric sequences, due to the (mis)incorporation of rNMP into DNA by DNA polymerase, is observed even in modern biology.1-2 And to repair such misincorporations, sophisticated proof-reading enzymes have been invented. But in prebiotic era there were no such sophisticated enzymes. Hence, the existence of such hybrid sequences (heterogeneity) in the early Earth would have been more realistic. So, what were the roles of those chimeric polymers in the evolution of homogeneous nucleic acids? Did they help or interfere, or were they bystanders, in the emergence of RNA and DNA?
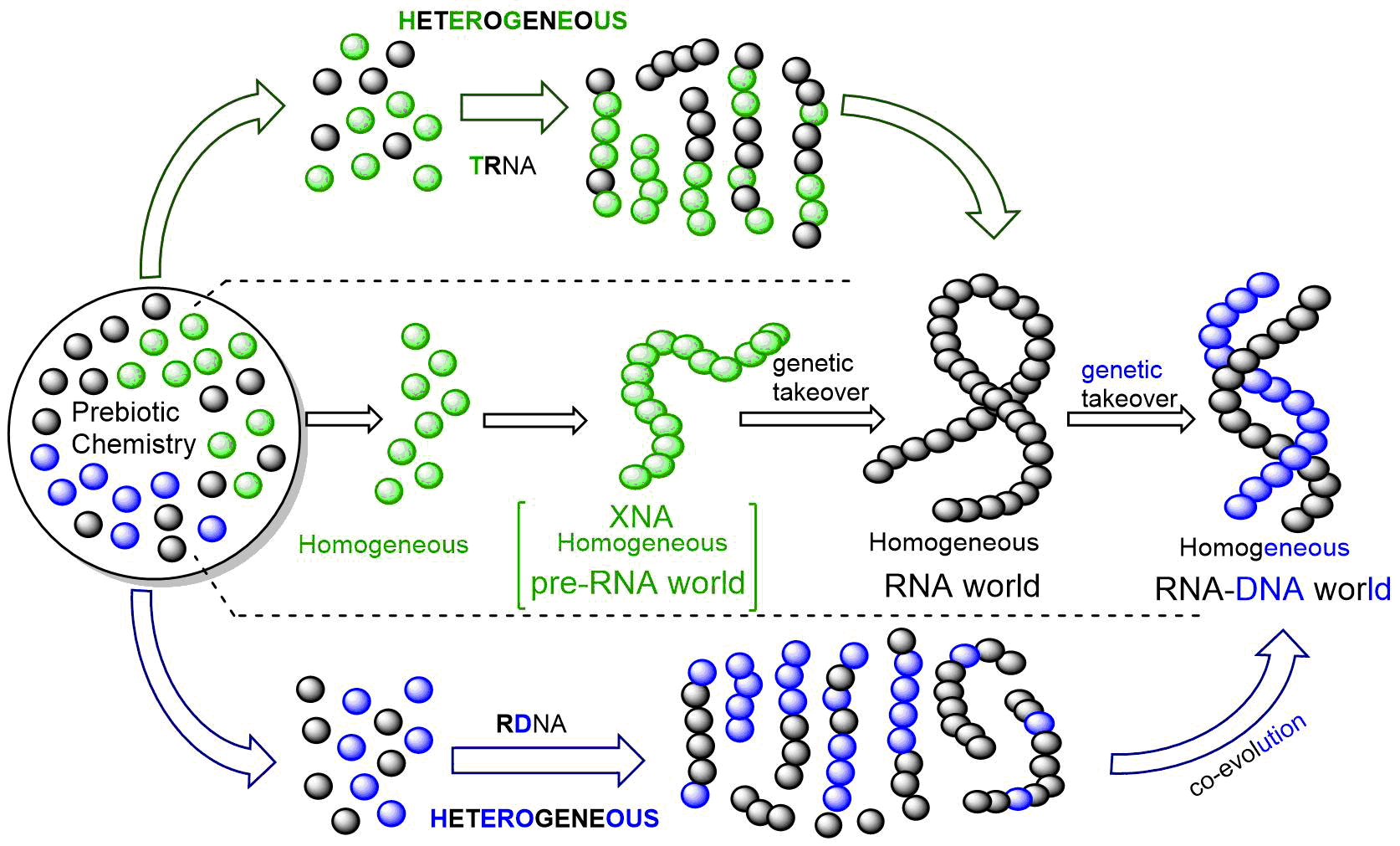
Figure-1: Scenarios for the emergence of RNA and DNA. The middle arrow indicates the popular hypothesis of the appearance of RNA from a prebiotic mixture, which later gives rise to DNA. The top and bottom arrows indicate about the possible emergence of RNA and DNA via hybrid sequences (heterogeneity to homogeneity).
To understand the role of such heterogeneity (chimeric sequences) we started our investigation by mixing TNA monomers into the backbone of RNA. We chose TNA as a plausible pre-RNA molecule as it is known to form stable Watson-Crick duplex3 and could be synthesized in the same way as RNA through the similar prebiotic routes4-5. After the discovery of TNA, Orgel speculated about a graduality scenario, where TNA is converted to RNA, by increasing substitution of RNA into TNA.6 As TNA cross-pairs with RNA, we anticipated that the mixing of TNA with RNA would also form stable chimeric-duplexes. But surprisingly, both in self-complementary and in non-self-complementary RNA sequences, such TNA insertion decreases the duplex-stability. Furthermore, an analysis of the base pairing results in self-complementary TNA-RNA hybrid sequences revealed an intriguing pattern (Figure 2). In self-complementary alternating TNA/RNA chimeric sequences (At)8 or (Au)8, the TNA-pyrimidine and RNA-purine combination formed stronger duplexes (Figure 2, left) when compared to the opposite combination of TNA-purine and RNA-pyrimidine i.e. (aT)8 or (aU)8. More surprisingly, this trend was reversed in block TNA/RNA chimeric sequences i.e. a8T8 or a8U8 where the TNA-purine and RNA-pyrimidine gave stronger duplexes over the inverse combination, t8A8 or u8A8 (Figure 2, right).
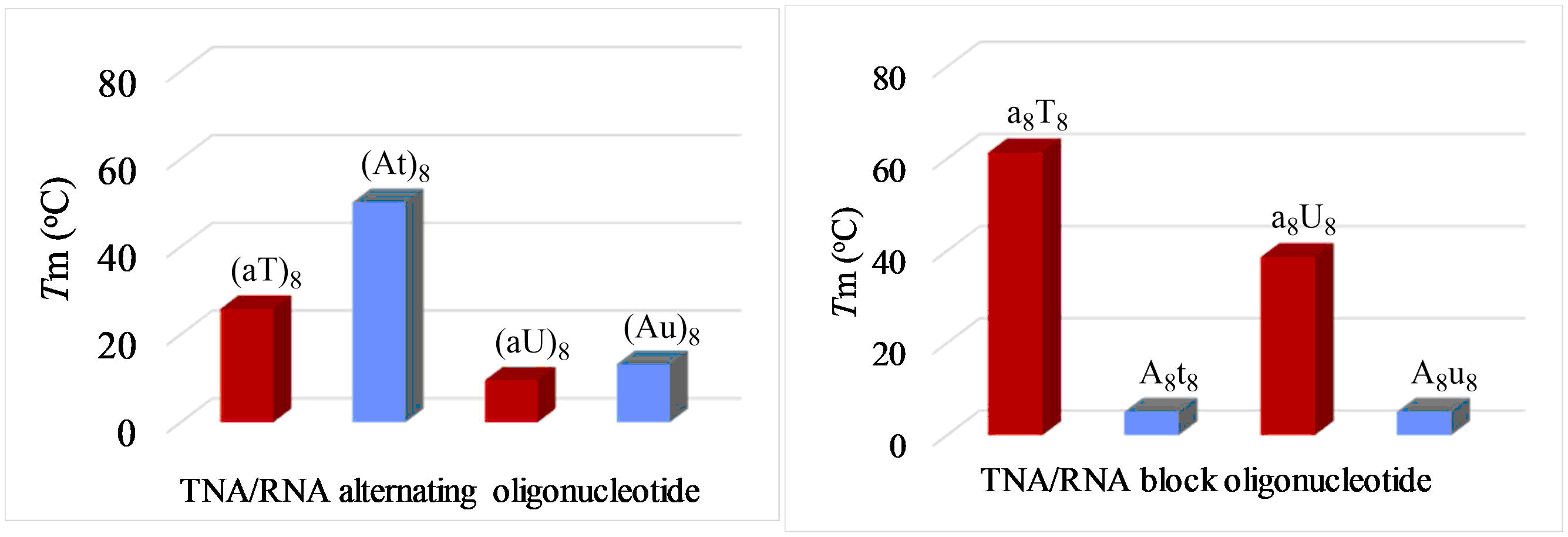
Figure. 2 Comparison of UV-Tm values of duplexes of alternating TNA-RNA chimeric sequences (left) and block TNA-RNA chimeric sequences (right) in 1M NaCl, 10 mM Na2HPO4, 100 µM EDTA of pH 7.2; a,t,u = TNA and A,T,U = RNA
These observations suggested that the non-self complementary sequences containing RNA-pyrimidine with TNA-purine in a block pattern could be combined with TNA-pyrimidine and RNA-purine at alternating positions may give rise to form strong chimeric base-pairing systems. Based on this hypothesis, two complementary strands were designed in such a way that the block portion of the chimeric sequence contains RNA(T) and TNA(a), while the alternating portion of the chimeric sequence contains TNA(t) and RNA(A).
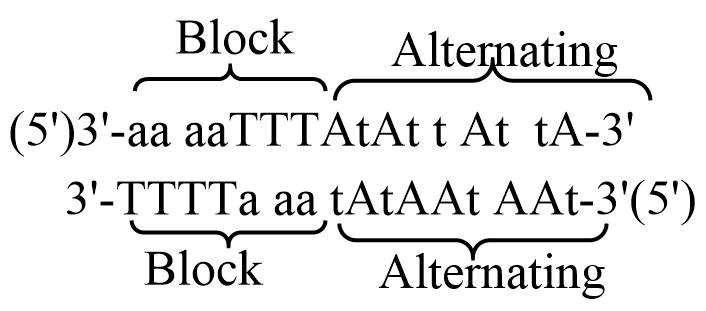
Figure 3. The complementary TNA-RNA chimeric sequences designed based on the strong-weak base pairing behavior as shown in figure 2. a,t = TNA; A,T = RNA.
Those two hybrid TNA-RNA sequences were synthesized, and their duplex stabilities were recorded. To our dismay, contrary to our predictions, they showed weak affinity for each other. We were initially devastated by this result; but then out of sheer curiosity, with no expectations, we checked whether these individual chimeric strands (as shown in fig. 3) would pair with their complementary homogeneous RNA strands. We were pleasantly surprised that the TNA-RNA hybrid strands did indeed form strong duplexes with RNA, and was found to be true for homogeneous DNA and TNA strands as well. I still can remember that day; it was on October 15th, 2015, and was the inflection point for this project. To confirm this unexpected behavior, we further carried out experiments with other TNA-RNA hybrid sequences and found that this behavior to be true and general for TRNA chimeric sequences containing all four nucleobases (A, T/U, G and C). Now the question arose, whether these chimeric sequences can act as templates in non-enzymatic template directed ligation reactions, because non-enzymatic template directed ligation is important for self-replication, which has been a stumbling block for the RNA world hypothesis due to the product-inhibition problem.7 To find out, we performed ligation reactions on TNA-RNA hybrid templates and found that they indeed ligate the homogeneous ligands more efficiently than the corresponding hybrid ligands. Such preference for homogeneous ligands was observed even when there is the mixture of both homogeneous and hybrid ligands, suggesting that homogeneous-backbone products are the favored outcome.
The question of whether the above selection of homogeneous-backbone sequences by hybrid-strands is limited to TRNA sequences or is general for other mixed-sugar-backbones, inspired us to investigate hybrid RNA-DNA sequences. The plausible coexistence and coevolution of RNA and DNA in prebiotic scenarios has been discussed before8-10. Accordingly, the base-pairing study with RDNA hybrid sequences were carried out and, to our delight, a similar preference for homogeneous-backbone sequences by hybrid RDNA template was observed and the rate of ligation reaction follows the order: homogeneous ligands-homogeneous template > heterogeneous ligands-homogeneous template ≈ homogeneous ligands-heterogeneous template>> heterogeneous ligands-heterogeneous template. Such preference of homogeneity-homogeneity and homogeneity-homogeneity over the heterogeneity-heterogeneity was further utilized, as described in the proof-of-principle study, for overcoming the primary limitation in the non-enzymatic template directed ligation reaction- the template-product strand inhibition where the continuous production of the product is curtailed due to the strong association of the initially formed template-product complex. Furthermore, we were able to demonstrate that chimeric sequences can also help in a cross-catalytic self-replication with potential for amplification.
For further details of the work in overcoming template-product inhibition and potential cross-catalytic self-replication click here. The work is published in Nature Chemistry.
References
- Joyce, C. M. Choosing the right sugar: How polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. U.S.A. 94, 1619-1622 (1997).
- Cerritelli, S. M. & Crouch, R. J. The balancing act of ribonucleotides in DNA. Trends Biochem. Sci. 41, 434-445 (2016).
- Schoning, K.-U. et al. Chemistry of alpha-aminonitriles. 37. The alpha-L-threofuranosyl-(3'->2')- oligonucleotide system ("TNA"): Synthesis and pairing properties. Helv. Chim. Acta 85, 4111-4153 (2002).
- Kim, H.-J. & Benner, S. A. Prebiotic stereoselective synthesis of purine and noncanonical pyrimidine nucleotide from nucleobases and phosphorylated carbohydrates. Proc. Nat. Acad. Sci. U.S.A. 114, 11315-11320 (2017).
- Islam, S. et al. Detection of potential TNA and RNA nucleoside precursors in a prebiotic mixture by pure shift diffusion-ordered NMR spectroscopy. Chem. Eur. J. 19, 4586-4595, (2013).
- Orgel, L. E. Perspectives: Origin of life: A simpler nucleic acid. Science 290, 1306-1307 (2000).
- Fernando, C., Von Kiedrowski, G. & Szathmáry, E. A Stochastic Model of Nonenzymatic Nucleic Acid Replication: “Elongators” Sequester Replicators. J. Mol. Evol. 64, 572-585, (2007).
- Orgel, L. E. & Lohrmann, R. Prebiotic chemistry and nucleic acid replication. Acc. Chem. Res. 7, 368- 377 (1974).
- Oró, J. & Stephen-Sherwood, E. The Prebiotic synthesis of oligonucleotides. In: Oró, J., Miller S.L., Ponnamperuma C., Young R.S. (eds) Cosmochemical Evolution and the Origins of Life. (Springer, Dordrecht, 1974).
- Sutherland, J. D. & Whitfield, J. N. Prebiotic chemistry: a bioorganic perspective. Tetrahedron 53, 11493-11527 (1997).
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in