Redefining the global expansion of a marine pathogen clone into Latin America with an eco-evolutionary approach
Published in Earth & Environment, Ecology & Evolution, and Microbiology

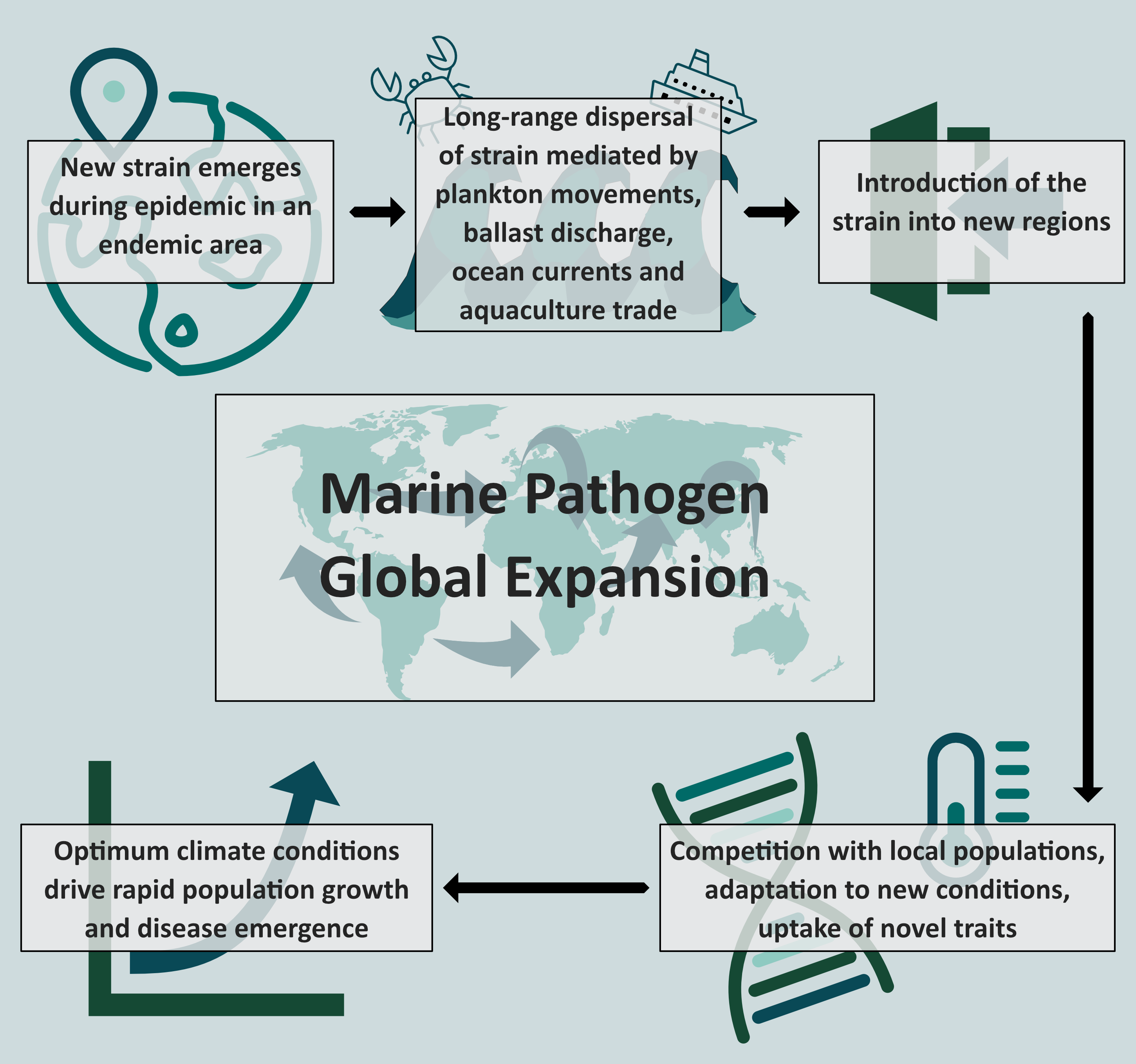
Global expansion of marine pathogens
Our knowledge of the biological diversity and ecology of marine microorganisms in the oceans has remained incomplete. It is difficult to obtain samples across vast areas of the oceans, and such samples often are of a limited resolution to describe community assemblage and intra-species variability. These constraints have also restricted our understanding of the spatial distribution and biogeographical patterns of microbial communities in the oceans and the routes of migration connecting populations between different oceanic regions.
These aspects are particularly relevant for marine pathogens like Vibrio, water-borne bacteria that cause illnesses such as gastroenteritis. Pathogenic Vibrio species (including V. cholerae and V. parahaemolyticus) are characterised by complex population structures with a mixture of genetic variants, comprising local strains alongside variants that emerge thousands of miles away from their areas of endemicity. Different routes and dispersal mechanisms have been suggested to explain this particular interconnectivity characteristic of Vibrio populations, ranging from release of ballast water transported by cargo ships and long-distance dispersal in ocean currents, however the open ocean is often described as too hostile an environment for Vibrio survival. Studying Vibrio and their associated diseases can be very challenging, with little evidence of the routes and mechanisms of this global spread and introduction of strains, making the global expansion of Vibrio one of the most indescribable phenomena across pathogenic organisms (Colwell, 1996).
Therefore, the global picture of Vibrio within the marine environment remains largely undefined. Specifically, understanding the evolutionary mechanisms driving rapid adaptation to climates that are completely distinct from their primary endemic region of tropical Asian waters is a fundamental element to define the epidemic potential of these pathogens and recognise the rate of success of introductions in different regions.
Unravelling the evolutionary history of a Vibrio parahaemolyticus clone
Our study aimed to provide insight into this global expansion into distinct marine climates. We specifically investigated the rapid emergence of the pandemic clone Vibrio parahaemolyticus Sequence Type 3 (VpST3) in Latin America, a region with one of the most dynamic and variable climates of the world as a consequence of the impact of the El Niño Southern Oscillation.
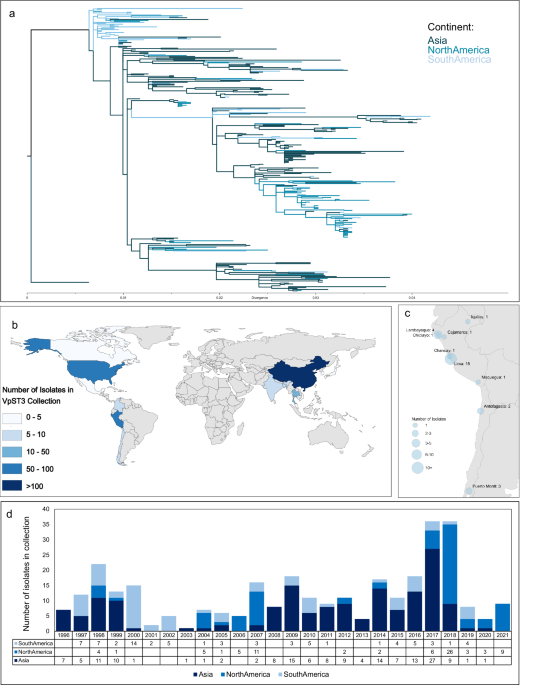
The emergence of VpST3 was first reported in association with a large epidemic in Calcutta, India in 1996 and underwent a rapid expansion through the Southeast Asian region over the following months. This strain was reported for the first time outside the Asian Continent in 1996, associated with infections in Peru, followed by a local propagation throughout South America, which was considered a spread of the ST3 strain that emerged in India.
Emergence of a well-adapted population in Latin America
However, our study reveals an evolutionary and epidemiological scenario completely different from that previously reported and accepted for this pathogen, decoupling the epidemic in Latin America from its emergence in Asia.
Using novel genomic isolates collected in Latin America from 1997 onwards, we identified a distinct population of VpST3 in Latin America. We found that the emergence of VpST3 in Peru was not a transcontinental spread of the epidemic strain that emerged in Calcutta in 1996, but rather the result of the introduction in South America of an ancestral variant of the VpST3 well before its emergence 1996, where it underwent a successful adaptation to local conditions over its evolutionary divergence from this ancestral stage, and from VpST3 strains found in Asia. We found selection signatures for genes that increased fitness and survival in the distinct marine climate of Latin America, providing resilience to the colder, more saline and more acidic waters off the coasts of Peru and Chile.
The story then becomes clearer: when this variant arrived in Latin America (such as through long-range dispersal mechanisms, potentially mediated by plankton migration due to particular attachment traits identified), these unique traits provided advantages during competition with other local populations, facilitating selection of this variant that had higher fitness in the new conditions upon arrival. Vibrio bacteria are opportunistic, and therefore as soon as optimum conditions returned (likely the exceptional warming conditions associated with the strong El Niño event in 1996-97), VpST3 underwent rapid population growth and propagated throughout Latin America, causing outbreaks in its wake, and resulting in this strain becoming dominant in Latin America. Such patterns, with outbreaks being identified before any previous records of these variants in an area, have been observed for a range of Vibrio parahaemolyticus clones across the world, such as ST36 in New Zealand, Spain and New York.
The new age of analysis: using genomic and environmental data in combination
This analysis was possible due to the application of an eco-evolutionary analytical framework, combining genomic data, evolutionary analysis and oceanic climate data in an inter-disciplinary approach. Using these datasets in combination has provided a novel perspective of the eco-evolutionary dynamics driving global expansion of pathogenic organisms and revealed novel insights into the biological mechanisms that enable to successfully colonize new regions with distinct marine environments.
Going forward, we will continue to characterise the mechanisms at the eco-evolutionary interface that drive the global expansion of marine pathogens, to facilitate forecasting of the impacts of resulting outbreaks and pandemics on human health, particularly in dynamic marine environments modulated by climate change.
The next chapter
Vibrio populations are constantly dynamic, identifying more efficient strategies to survive in different environments, exploring alternative evolutionary pathways. As part of this journey, a new variant has emerged with stronger adaptive advantages which appear to have overpowered this Latin American variant, with this new variant becoming dominant both in Peru and around the world. But this is a different story, and represents a new chapter in the dynamic and exciting evolution of VpST3.
Read the full story here: Evolutionary dynamics of the successful expansion of pandemic Vibrio parahaemolyticus ST3 in Latin America | Nature Communications.
References
Colwell, R R. “Global climate and infectious disease: the cholera paradigm.” Science. 274, 5295 (1996)
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in