Sensing low osmotic pressure to produce flagella
Published in Microbiology

Many pathogenic enterobacteria have flagella which enable them to move towards favorable environment such as high nutrient concentration. In contrast, Escherichia albertii, a close relative of Escherichia coli, had been reported non-motile and non-flagellated although it carries flagella-related genes. When we tried to take their images by scanning electron microscopy (SEM), we incidentally found a few flagella on their surface.
Initially, we fixed the bacterial cell culture on cover glass for preparation of SEM samples, but many debris were scattered on the samples. To reduce the number of the debris, we used diluted medium to grow bacteria and it worked. When we looked at the newly acquired images, we were surprised because there were structures that E. albertii would not have: flagella. We realized that the strains which produce flagella in diluted medium do not have flagella in ordinary medium. These findings prompted us to study flagella and motility in E. albertii.
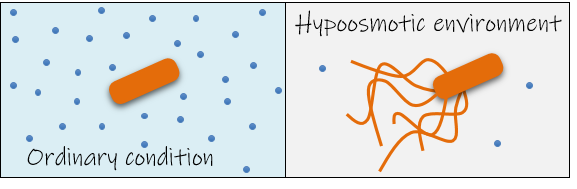
We tested various culture media to determine whether specific components of culture medium are responsible for motility induction in E. albertii cells. Surprisingly, E. albertii cells responded to restriction of salts to start swimming. We eventually found out that low osmotic pressure induces flagella synthesis in E. albertii. To our knowledge, this is the first report demonstrating that flagellar synthesis is induced by hypoosmotic stress.
E. albertii is a pathogen causing diarrhea and gastroenteritis in humans. Since flagella on E. albertii almost disappeared at osmotic pressures as high as that of human plasma, we asked whether flagella affect virulence of E. albertii. Flagella-induced and non-induced E. albertii cells were infected to human intestinal epithelial cells and their survival in the presence of antibiotics was evaluated. The number of surviving cells in flagella-induced E. albertii cells was 130-fold that of non-induced cells, indicating that the flagella promote invasion in intestinal epithelial cells, thereby increasing bacterial survival in the presence of antibiotics.
Meanwhile, we collaborated with Dr. Tadasuke Ooka, Kagoshima University, Japan, who studies genome structure of E. albertii. A total of 59 E. albertii strains were examined for motility and 27 (45.8%) of them showed swimming ability in the diluted medium. The analysis of whole-genome sequencing data revealed that the motile E. albertii strains have a set of flagella-related genes while the non-motile strains have mutations on these genes.
Flagella are energetically expensive to synthesize and operate but E. albertii does not carry any chemotaxis-related genes required for the directed, active movement towards or away from specific chemical substances. What does it mean for bacteria to swim freely? E. albertii would become a useful tool for studying motility and its directional control.
Read the full paper at:
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in