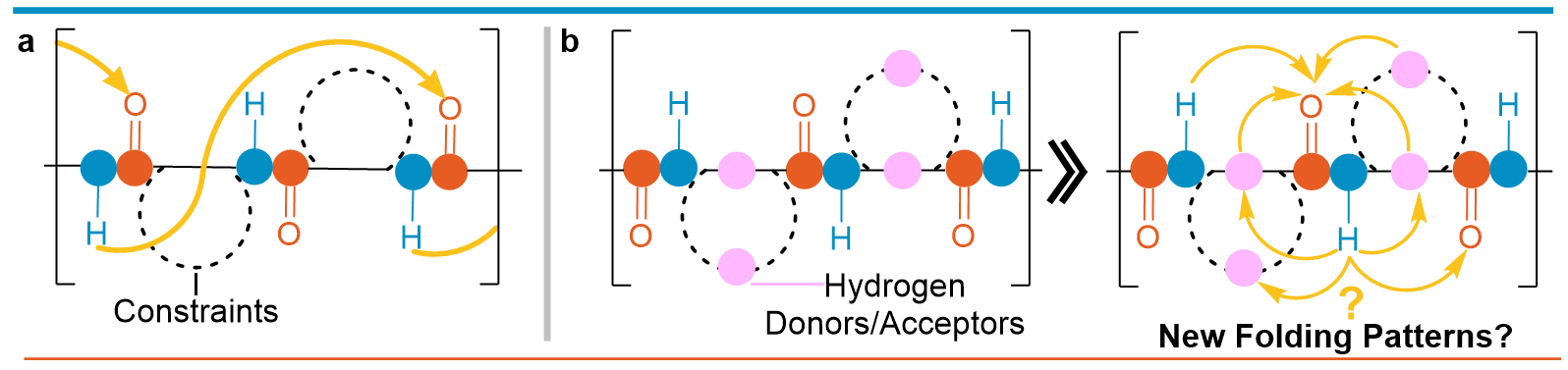
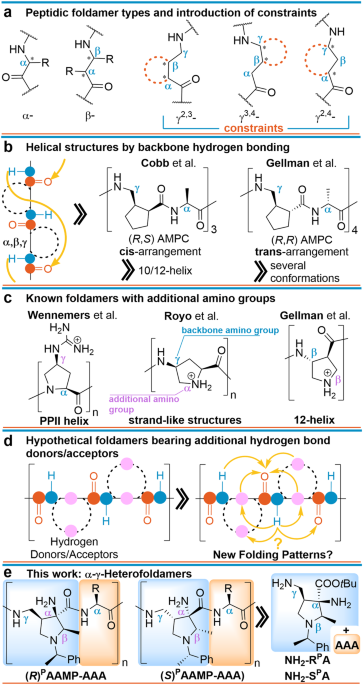
The precise arrangement of large biological molecules is vital for life's processes. Proteins, composed of L-α-amino acids, perform their functions through specific structures, such as β-strands and α-helices. Mimicking these natural structures, researchers study foldamers - artificially ordered systems. Among these, peptide-like chains containing constrained non-natural amino acids are popular for their diverse structures and potential applications in materials, drug delivery, and catalysis. In these areas, research on foldamers mainly focuses on understanding how specific structural constraints, backbone stereochemistry, and intramolecular hydrogen bonding influence their secondary structural shapes (Figure 1a). However, most non-natural foldamers adopt relatively rigid structures in nonpolar solvents, while their ability to fold in aqueous environments is rather rare.
Considering the introduction of extra hydrophilic functional substituents, the backbone may gain new interactions and organize itself to unique folding patterns potentially making them compatible with aqueous environments. Conversely, nature demonstrates that the significant presence of charged and hydrophilic amino acids capable of forming hydrogen bond networks may increase their mobility resulting in rapidly interconverting conformations, as seen in intrinsically disordered proteins. Guided by these facts, we hypothesize that non-natural amino acids, which provide additional free amino groups, both at the periphery and along the backbone may significantly affect the folding patterns and result in unique secondary structure motifs (Figure 1b).
Several years ago, we developed a concise two-step method accessing (2R,3R,4R)-1-((R)-1-phenylethyl)-3-amino-4-(aminomethyl)-2-methylpyrrolidine-3-carboxylates ((R)PAAMP) NH2-RPA from readily available materials, via tandem reaction and reductive cleavage of a diazo group on resulting pyrrazolopyrrolidine 1 (Figure 2).1,2 Conveniently, this polyfunctional amino acid could be orthogonally tuned to be compatible with Fmoc/tBu peptide synthesis strategy, and the protocol enables preparation of both enantiomers, NH2-RPA and NH2-SPA. We supposed that peptide composed of NH2-RPA and glycine would form a basic structural pattern. Replacing glycine with L-alanine would either stabilize or destabilize this pattern depending on its match with enantiomer NH2-RPA or NH2-SPA. At this point, we sought to thoroughly investigate the structural behavior of peptides incorporating all combinations of these polyfunctional building blocks with either glycine or L-alanine. Our assessment proved correct, and we were able to see consistent behavior of Pg-GRPA and Pg-ASPA series, while Pg-ARPA exhibits distinct spectral and physical features, including irreversible formation of insoluble aggregates.
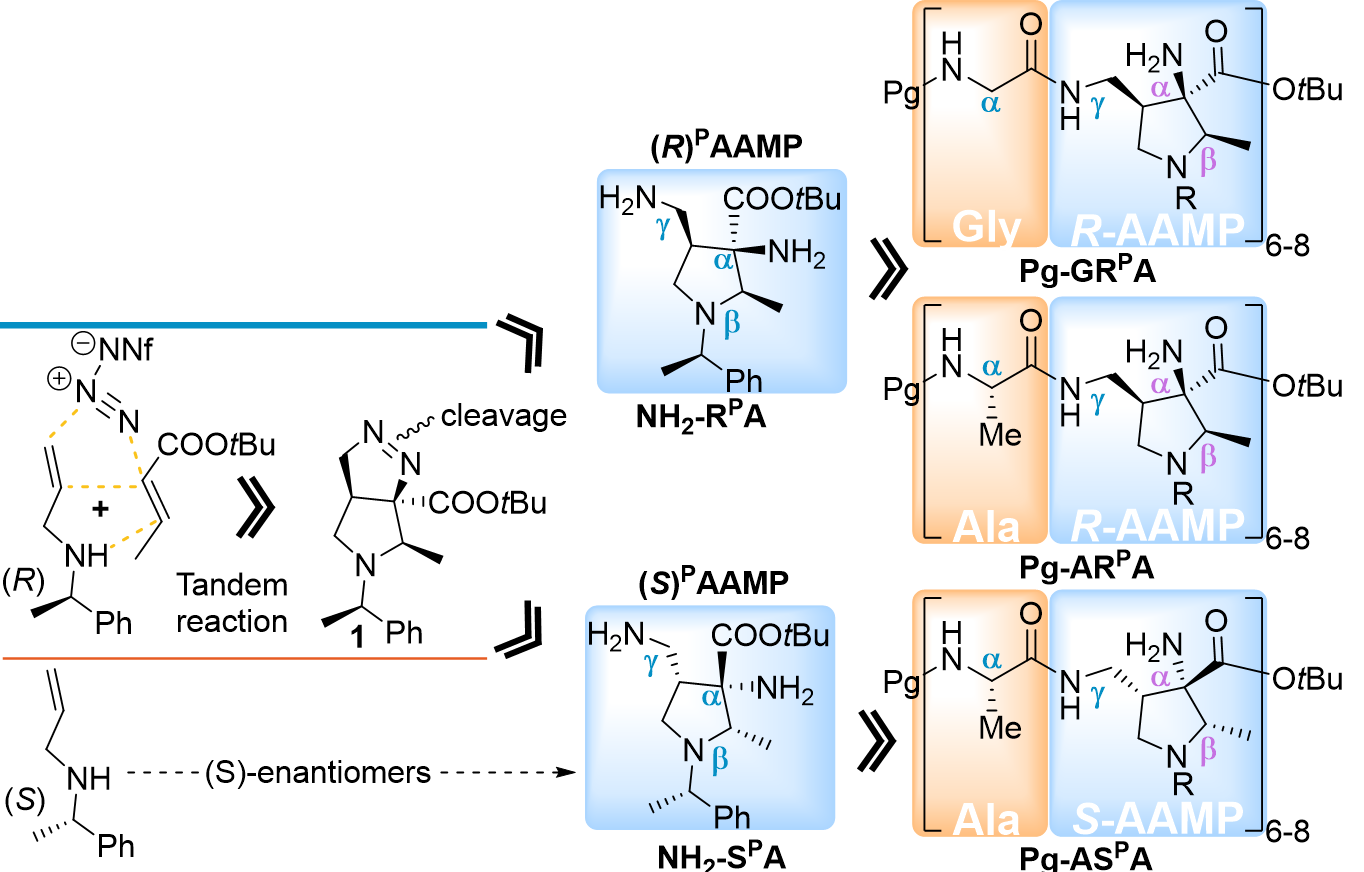
Figure 2. The original methodology behind this work and depiction of the three investigated series.
We aimed to investigate the identity of secondary structure. The first piece of the puzzle was obtained by the X-ray crystal structure of tetrapeptide NH2-4GRPA, which revealed an NH···NH2···NR3 hydrogen bond network (Figure 3). This network bridges the AAMP unit via two five-membered rings, locking its backbone geometry in an unprecedented triazaspiranoid-like arrangement. Next, we conducted NMR studies, including titrations and ROESY experiments, which confirmed the presence of these hydrogen bond networks in all higher oligomers in solution. We also identified long-range NOE interactions, providing an initial idea of the overall structural shape. To obtain a complete structural picture, we combined the NMR experimental results with molecular dynamics simulations and quantum chemical calculations. Molecular dynamics simulations indicated preferential occupation of two secondary structures: an 18/20 right-handed helix and a left-handed staircase-like structure. Finally, we leveraged chiroptical methods, including ECD and VCD, which confirmed the right-handed helical arrangement and also showcased its presence in the aqueous environment.
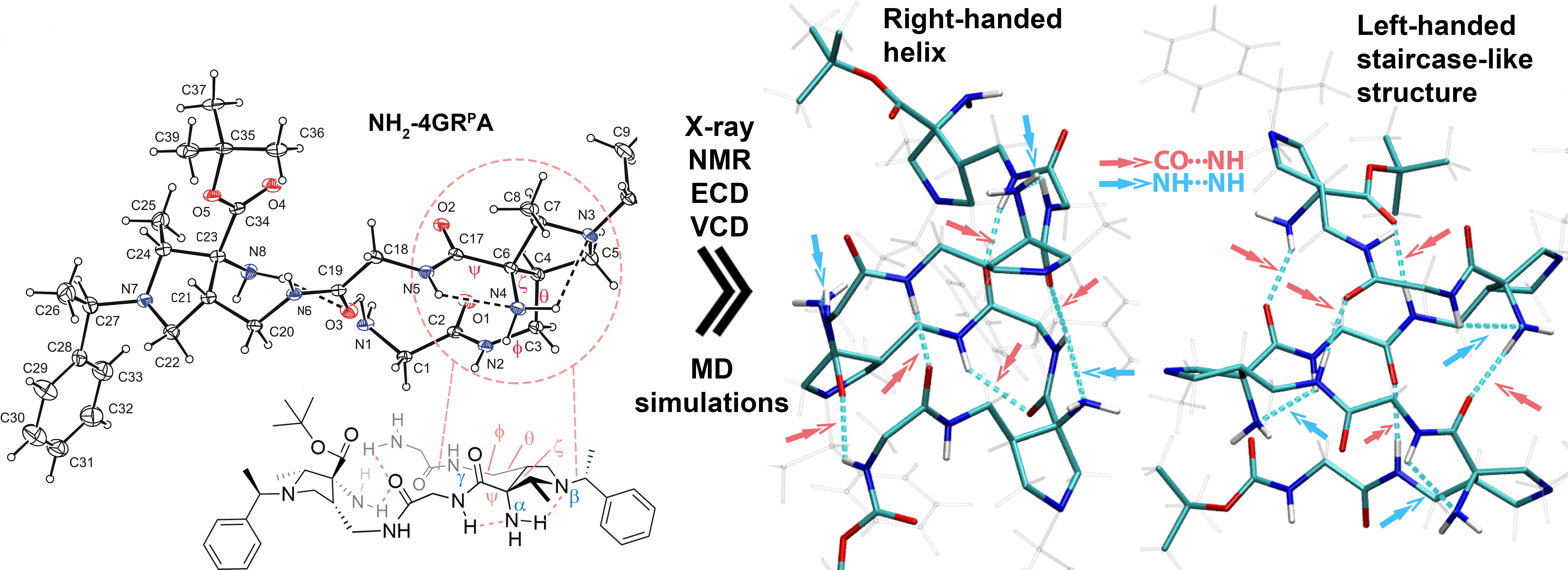
Figure 3. Secondary structure elucidation. X-ray crystal structure of NH2-4GRPA with double five-membered hydrogen bond as a crucial structure-forming element and visualization of computed representative right-handed helical and left-handed staircase-like secondary structures.
Our study confirms that additional amino groups dominate the hydrogen-bonding pattern through NH···NH2···NR3 arrangements, playing a crucial role analogous to the common peptide amide backbone hydrogen bonds and conformational constraints in shaping the secondary structure. Similar structures were found in peptides with both flexible glycine and helix-inducing alanine units. Thus, incorporating AAMP units with various natural amino acids may achieve wide helical arrangements. In general, this concept of involving extra functional groups with hydrogen-bonding capabilities may enhance specific folding propensities in peptides, opening new directions in the engineering of bio-inspired materials.
References
- Kapras, V., Pohl, R., Císařová, I. & Jahn, U. Asymmetric Domino Aza-Michael Addition/[3 + 2] Cycloaddition Reactions as a Versatile Approach to α,β,γ-Triamino Acid Derivatives. Org. Lett. 16, 1088–1091 (2014).
- Just, D., Hernandez-Guerra, D., Kritsch, S., Pohl, R., Císařová, I., Jones, P. G., Mackman, R., Bahador, G. & Jahn, U. Lithium Chloride Catalyzed Asymmetric Domino Aza-Michael Addition/[3 + 2] Cycloaddition Reactions for the Synthesis of Spiro- and Bicyclic α,β,γ-Triamino Acid Derivatives. Eur. J. Org. Chem. 5213–5221 (2018).
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Advances in Asymmetric Catalysis for Organic Chemistry
Publishing Model: Open Access
Deadline: Mar 31, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in