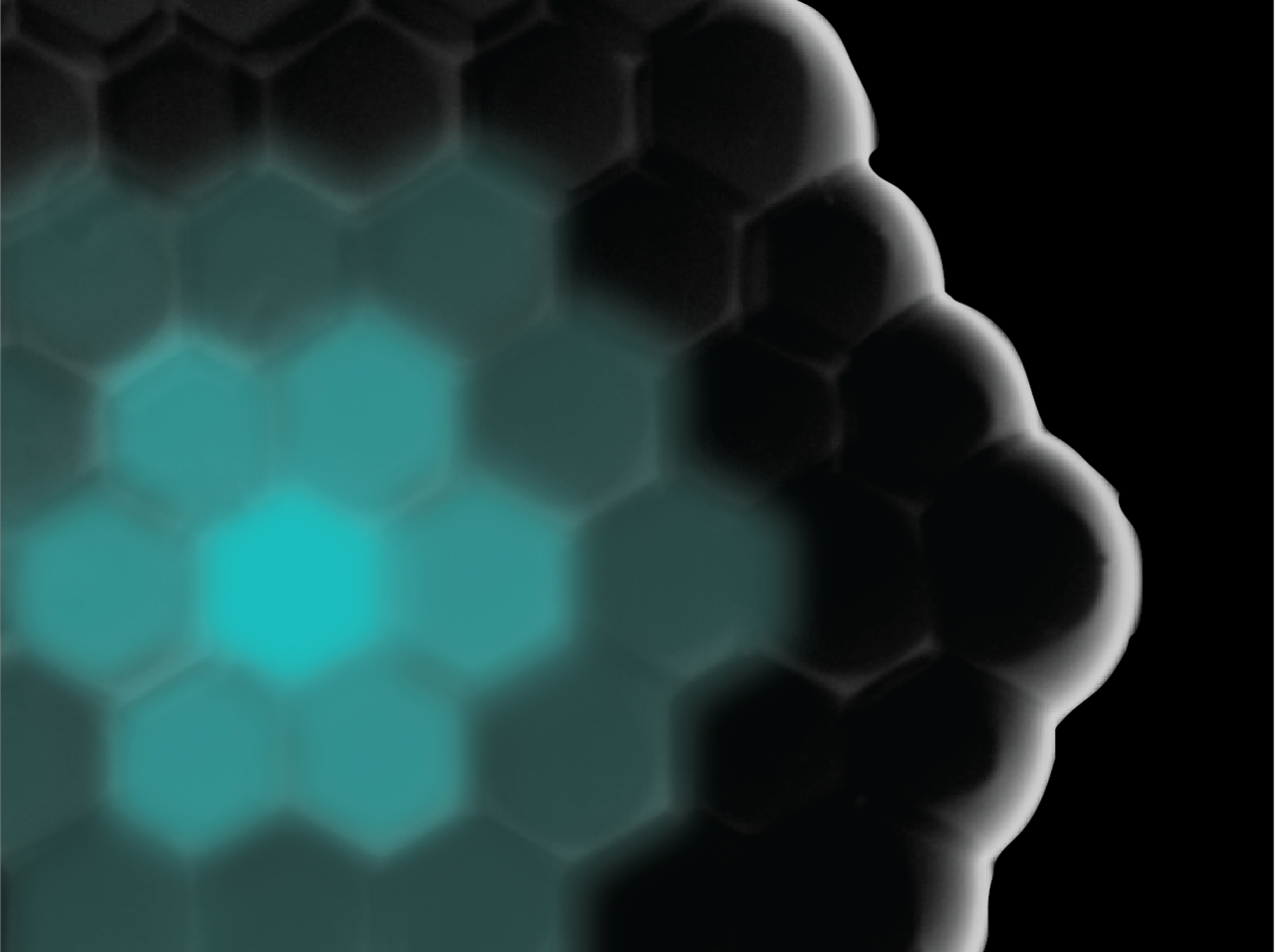
When I joined the lab of Prof. Simmel in 2014, we were interested in engineering artificial cells to reconstitute fundamental processes that may lead to emergent complex behavior and self-organization. Inspired by existing work in the field, I started with perhaps the simplest model for an artificial cell: a droplet of water in oil. Using the droplet-interface-bilayer technique, we brought single droplets into contact so that lipid bilayers – akin to real cell membranes – would form between the droplets.
Our first questions when we studied this system were quite simple: we wanted to know what could pass through this bilayer, and how specific the permeation is. We studied whether protein pores can form channels through the bilayer and facilitate chemical diffusion across it. We wanted to know if the chemical and physical properties of a molecule could predict how it propagated from droplet to droplet.
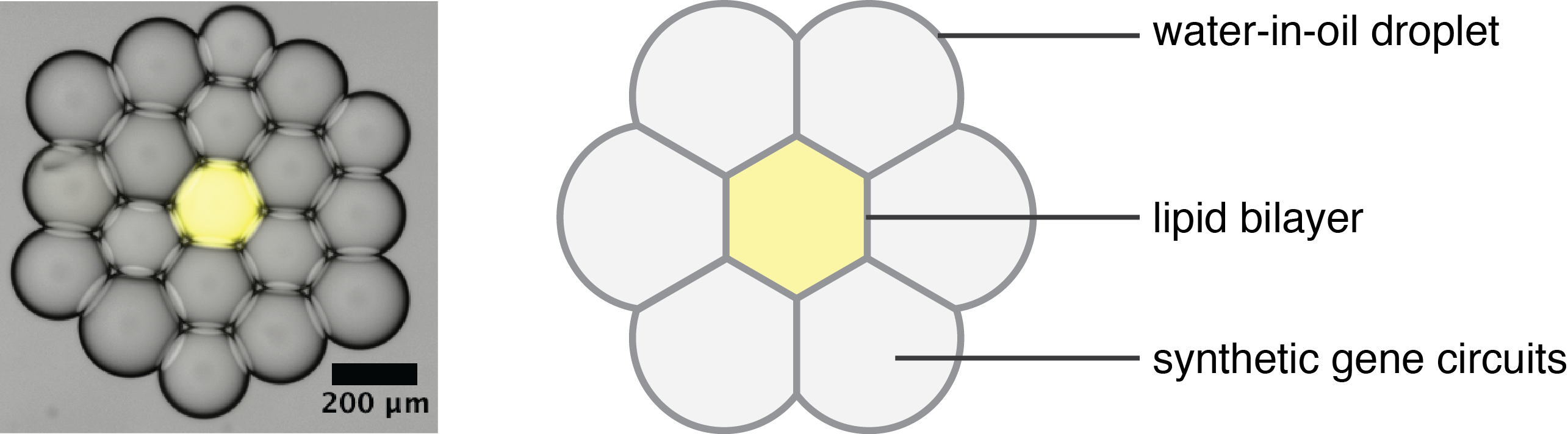
We eventually realized that the droplet-interface-bilayer technique provided us with control over the spatial organization of the networks, and therefore over spatial information. As the droplets can be brought in geometrical arrangements where the exact content and position of each droplet is perfectly controllable. Because of the protein pores, the communication between each compartment and its neighbors can also be controlled.
Most of the studies on engineered artificial cells focus on non-organized or randomly organized compartments, which resemble to a certain degree how unicellular organisms might be organized and interacting with one another. Our system, however, more resembles a multicellular tissue in some of its features, such as direct cell-to-cell communication, the possibility to establish overall chemical gradients, or the overall spatial cohesion. It gave us a great opportunity to investigate how complex behavior can arise in such spatially organized systems from simple chemical information. We therefore sought out to engineer synthetic biological circuits that could respond to spatial information.
We focused on a library of chemicals that could mediate communication by inducing synthetic circuits and sorted those that needed protein pores to accelerate their diffusion from those that didn’t. We modeled and measured how different processes of diffusion would affect downstream response genes. Then, we designed an in vitro DNA/RNA circuit based on a known gene network motif in living cells, the incoherent feed-forward loop 1. With this we were able to produce a propagating signaling pulse from a source droplet through an array. The velocity of this pulse could be tuned by internal and external parameters, and was affected by the geometry of the droplets network.
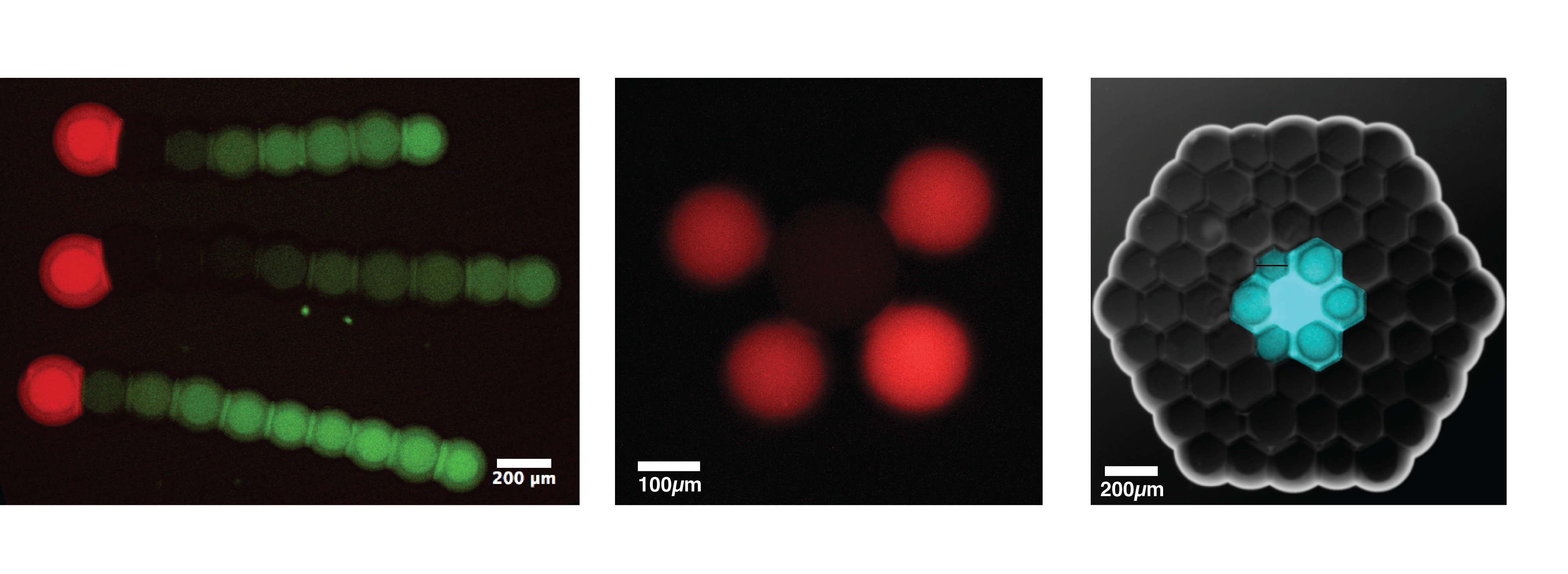
Embryogenesis is a particularly fascinating example of complex behavior emerging from noisy gene expression or from initially simple chemical processes that direct cell fate. To investigate if this is something we could reproduce in our systems, we engineered a circuit with a positive feedback loop that affected the permeability of the bilayers to a protein expression inducer. The droplets were loaded with E. coli cell extract and the circuit’s positive feedback loop was induced by noisy, leaky expression of the genes. We were able to measure significant differentiation in the protein expression levels between different droplets within the same network and attribute it to the stochasticity of key processes such as pores incorporation in the bilayer.
Our work suggests that providing artificial cell systems with spatial organization allows for complex emergent behavior, which opens new possibilities for the engineering of artificial cells. We think our set-up is a great platform to investigate fundamental processes, and we are particularly interested in mimicking dynamics and gene networks involved in embryogenesis.
Our paper can be found here: "Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits" in Nature Chemistry.
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in