
Imagine if everyone you met in life came with a label detailing each of their ex-partners? That may (or may not) be useful, but in the case of identifying which binding partners a protein has interacted with during its lifetime, this would be very valuable information.
The proteins in our bodies rarely act alone. In fact, almost every biological process that occurs within our cells is regulated by a complex network of interacting proteins. Protein-protein interactions (PPIs) are not just random events (i.e. proteins bumping into each other); their interacting surfaces have distinct topographies that fit together in a specific manner for particular purposes. For example, stable PPIs are found in multi-subunit complexes such as the proteasome, which is responsible for degrading damaged and unwanted cellular proteins. PPIs can also occur transiently, for example, in signalling pathways that control whether a cell lives or dies, while defective interactions can occur in disease states such as cancer. If novel diagnostics and targeted therapeutics are to be developed, we must have a thorough understanding of these interaction networks. For that reason, it is important that we have good tools available to study the interactions proteins make with each other.
We had the idea of taking a small section of a protein (a peptide) that mimicked a key interacting surface of the parent protein, and attaching a reactive group that would result in labelling of interacting protein partners. The labelled protein could then be identified following a specific readout. Our lab initially explored incorporating different types of reactive groups, but ultimately settled on using a peptide modified with Ru(II)(bpy)3, which can act as a light-activated single-electron transfer (SET) catalyst (Fig. 1). As a model system to develop our labelling approach, we chose the interaction between two B-cell lymphoma 2 (BCL-2) family members, MCL‑1 and NOXA‑B. Using sub-stoichiometric quantities of a Ru(II)(bpy)3-modified NOXA-B peptide, and irradiation with visible light for 1 minute, the anti-apoptotic protein MCL‑1 was labelled with a variety of functionalised dimethylaniline tags. Interestingly, we found that a cysteine residue on the protein had been modified with the label (in contrast to other reports on this type of SET-mediated protein labelling that reported tyrosine reactivity). We could also label MCL-1 selectively, in a mixture with other proteins, including the structurally related BCL-2 member, BCL-xL.
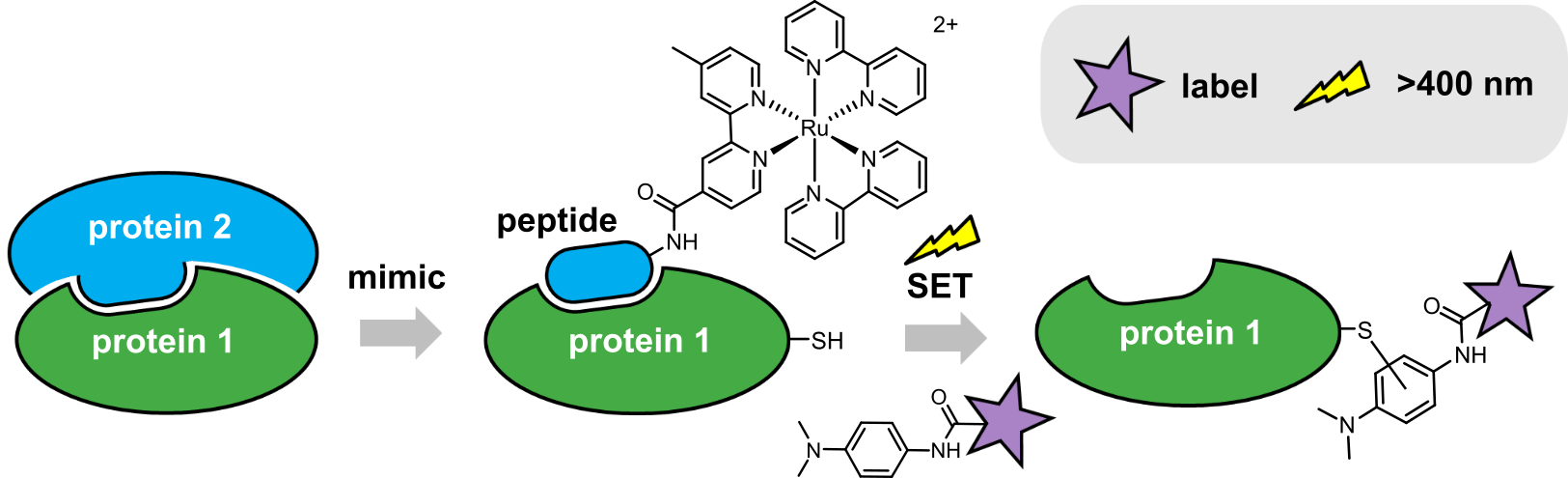
Figure 1: Ru(II)(bpy)3-mediated photolabelling of a protein using a peptide to mimic an interacting partner within a PPI, via a single-electron transfer (SET) mechanism.
The major advantages of this approach stem from the use of visible light to induce target protein labelling. Firstly, the Ru(II)(bpy)3-modified peptide and dimethylaniline label are unreactive in biological media, allowing equilibration to be reached prior to initiation of label transfer (reducing background labelling and consumption of the reagents by hydrolysis/reaction with bulk nucleophiles in the cell). Secondly, visible wavelengths are minimally damaging to proteins, orthogonal to other photoaffinity groups (such as diazirines) and can be achieved with simple and inexpensive equipment. Finally, temporal control over labelling may allow dynamic interaction studies: irradiation at specific time points to trap different protein complexes. Future studies using these types of reagents in cells may enable identification of novel transient and/or weak PPIs; presenting potential points for therapeutic intervention.
The full paper can be found here: https://www.nature.com/articles/s42004-019-0235-z
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Advances in Asymmetric Catalysis for Organic Chemistry
Publishing Model: Open Access
Deadline: Mar 31, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in