To See Ice in a Drop of Water
Published in Chemistry
It may not seem obvious at first glance, but our work was inspired by the “Auguries of Innocence” by William Blake:
“To see a World in a Grain of Sand”.
To some extent, most of computational chemistry is indeed describing the world based on a tiny model system of just a few hundreds of atoms. But here we mean something different: we found that all the local atomic environments observed in diverse ice phases can be found in liquid water. Or, to put it more poetically, we managed to see ice in a drop of water.
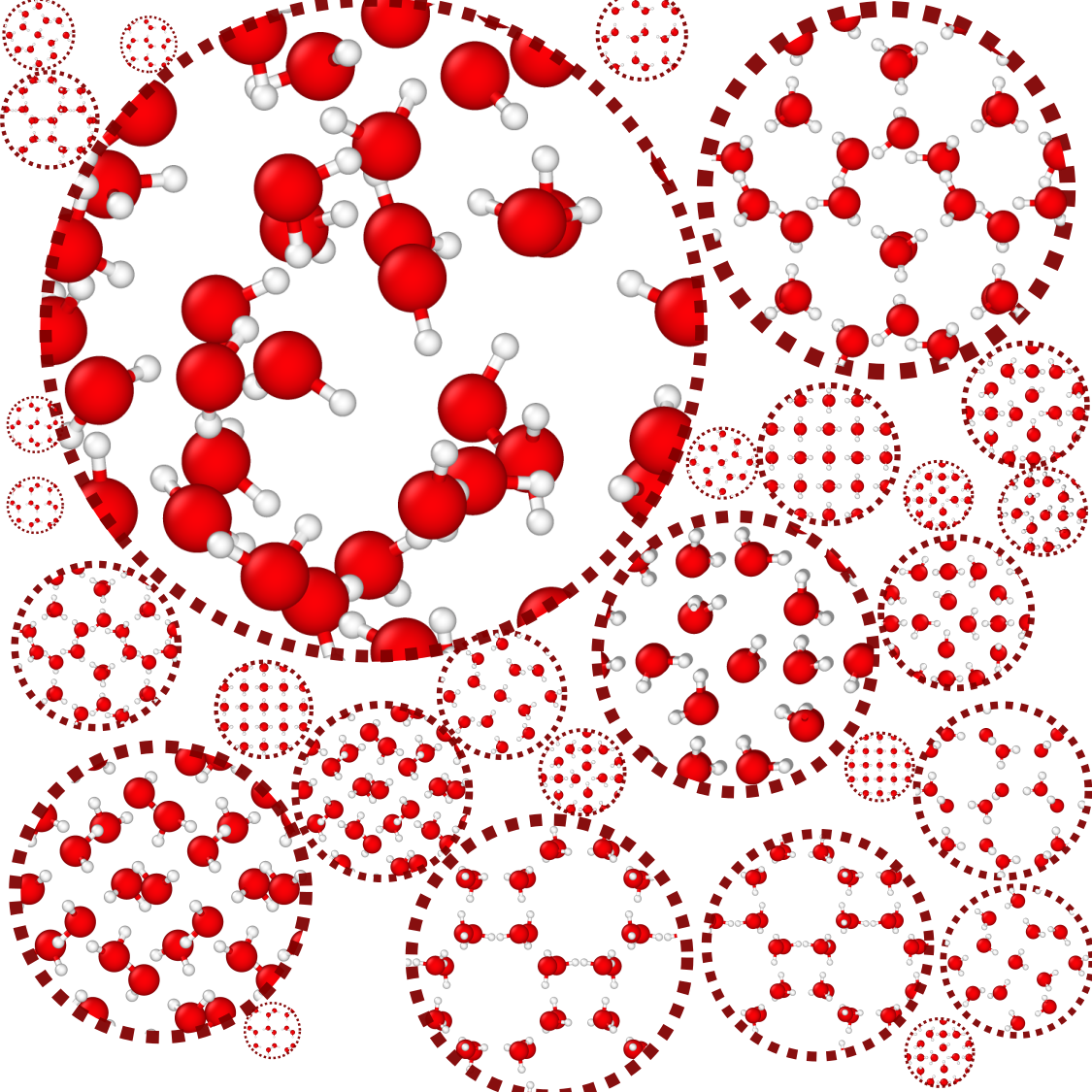
First of all, what is a local atomic environment in water? In our study, it is defined as all the neighboring atoms within a radius of a few Angstroms around a center of either oxygen or hydrogen, extending slightly beyond the second hydration shell of water molecules. We considered the local environments in liquid as well as in a total of 54 observed and hypothetical ice phases exhibiting enormous structural diversity. Several selected local environments in these phases are illustrated in Figure 1.
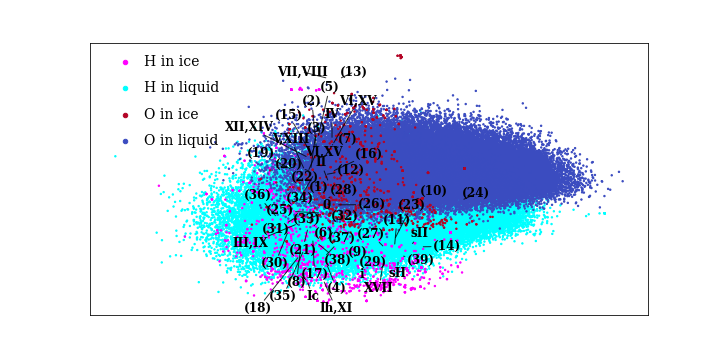
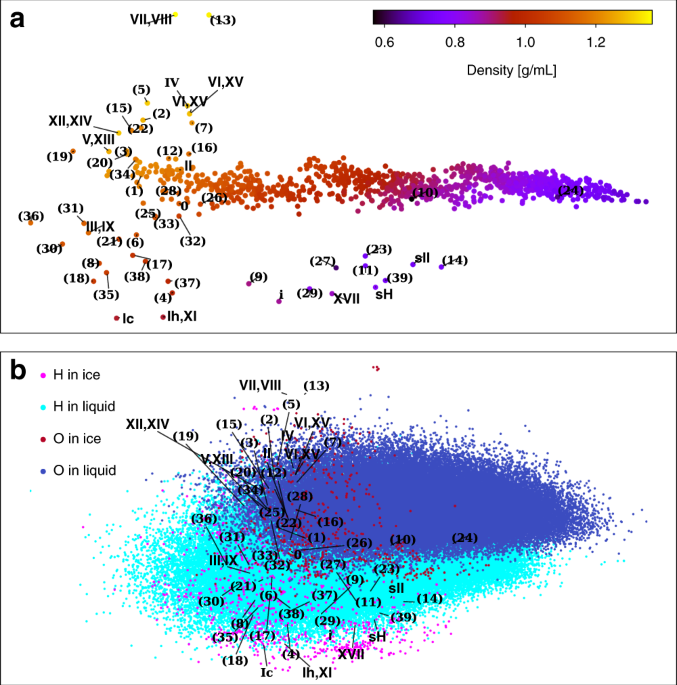
We adopted two strategies to systematically compare the local environments in the different ices and the liquid. The first is to directly compare the local environments using general atomic descriptors followed by a dimensionality reduction. In Figure 2, each dot represents the projection of each atomic environment, and it can be seen that the comparatively few distinct atomic environments in ices are almost completely covered by the continuum of environments realised in liquid water. Second, we demonstrate that a machine-learning potential (MLP) [1] that is only trained on liquid water accurately reproduces multiple ice properties including energies, densities, and how atoms vibrate inside the crystals. This means that the MLP can infer how the various ice phases behave by only learning about the liquid, because all ice local environments are present in the liquid.
We are, still to this day, amazed by how accurate the MLP is for describing ice phases that it has never seen before. From a practical point of view, this also implies that we can speed up the computation by a millionfold: for an ice structure, one can compute its vibrational properties on a laptop within a few minutes using the MLP, rather than the 16,000 CPU hours that we unfortunately had to spend using brute-force quantum-mechanical calculations. Besides such practical implications, the “ice in water” idea not only provides a new perspective on the understanding of water, but is also a humble tribute to William Blake.
References:
[1] Bingqing Cheng, Edgar A Engel, Jörg Behler, Christoph Dellago, Michele Ceriotti. (2019) ab initio thermodynamics of liquid and solid water. Proceedings of the National Academy of Sciences, 116 (4), 1110-1115.
[2] Bartomeu Monserrat, Jan Gerit Brandenburg, Edgar A. Engel, Bingqing Cheng. (2020) Liquid water contains the building blocks of diverse ice phases. Nat Commun 11, 5757. https://doi.org/10.1038/s41467-020-19606-y
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in