Uncovering abalone's genetic armour against heat stress
Published in Earth & Environment, Sustainability, and Genetics & Genomics
The Journey Behind the Research
Abalone, marine gastropods of the genus Haliotis, are ecologically vital, culturally significant, and prized as a premium seafood delicacy. Unfortunately, over 40% of abalone species are currently threatened with extinction. Driven by climate change and overexploitation, both wild and farmed populations are facing alarming declines. Understanding the genetic mechanisms underlying heat stress responses is essential for conserving these remarkable organisms and ensuring the long-term sustainability of abalone aquaculture.
Heat stress is the most widely studied environmental challenge in abalone research, and for good reason. As marine heatwaves become more frequent and intense due to climate change, both wild and farmed abalone populations are increasingly at risk. In response, researchers around the world have been using transcriptomic tools to understand how abalone react at the molecular level when exposed to high temperatures.
Over the years, numerous studies have investigated these responses in various species, under diverse conditions, and at different life stages. However, most of these studies were done independently, each providing only a piece of the puzzle. We realized this growing collection of data represented a unique opportunity: what if we could bring them all together and look for the common genes? In other words, are there core genes or pathways that are consistently activated when abalones face thermal stress, regardless of species, stress intensity or any other experimental factor (other than heat stress)?
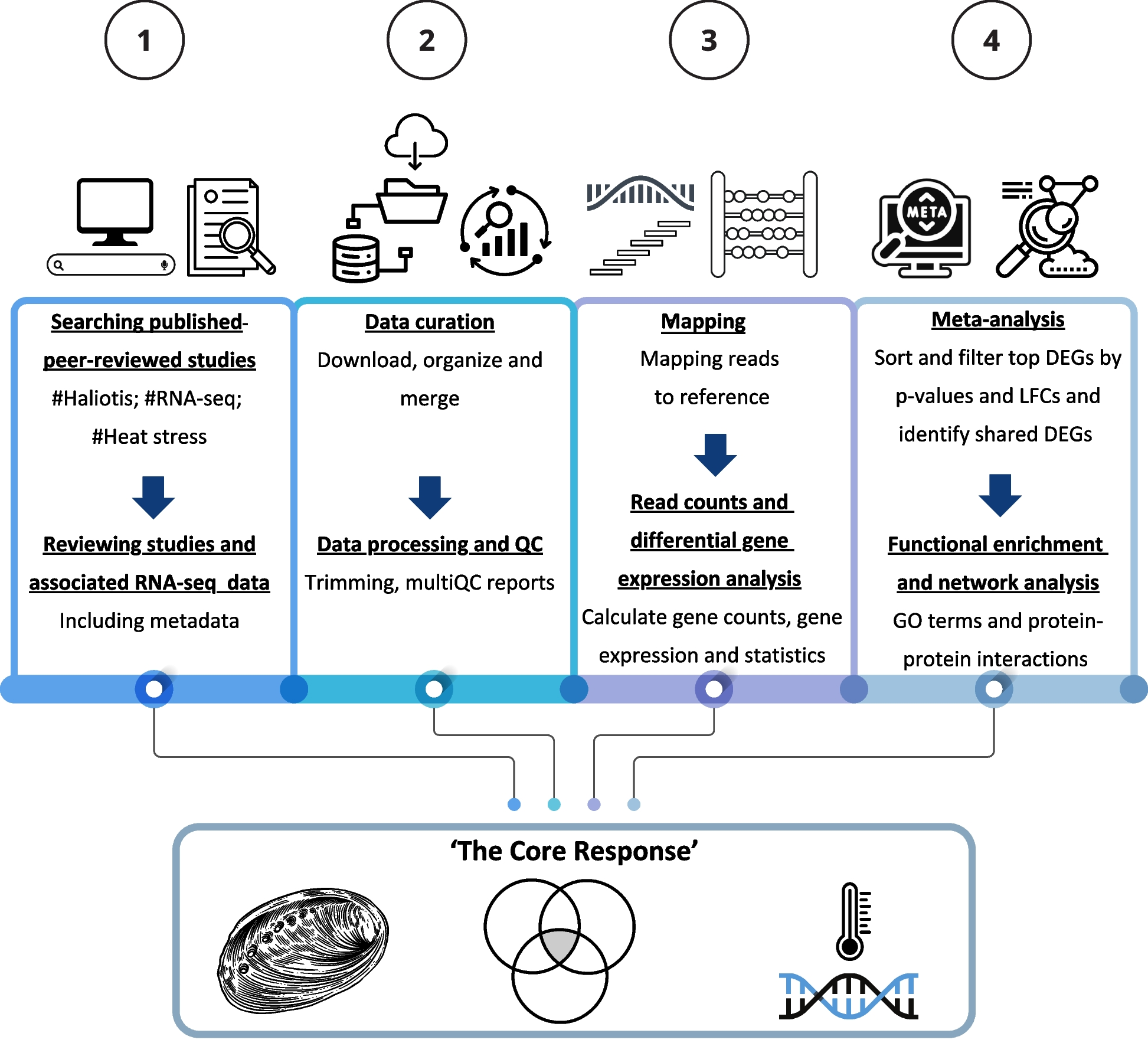
To answer this, we conducted a large-scale meta-analysis of publicly available RNA-seq datasets. Our goal was to integrate and reanalyze these studies through a single, consistent pipeline to uncover shared genetic patterns. Ultimately, we hoped to identify the key molecular players that help abalone survive in warming oceans and provide insights that could support climate-resilient aquaculture in the future.
Key Findings
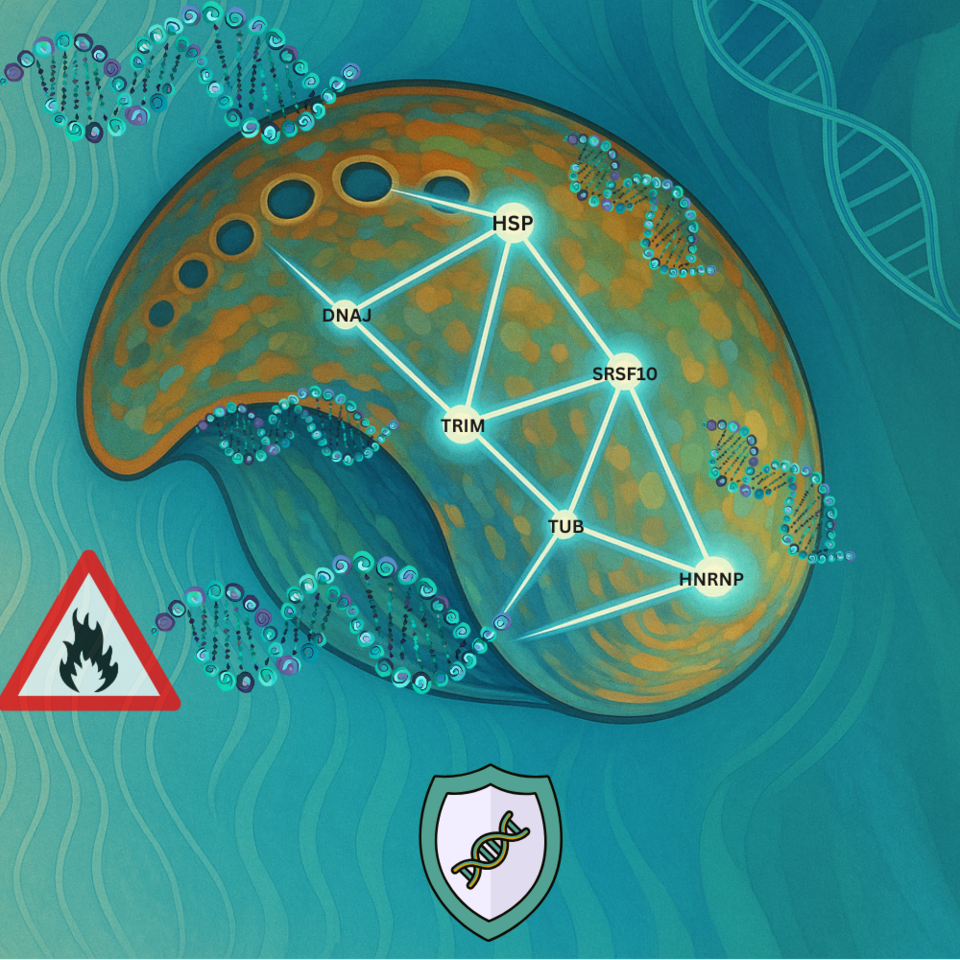
Through rigorous meta-analysis, we identified a remarkable pattern: a core set of 74 genes consistently responded to heat stress across most of the studies. These genes weren’t random as they formed a cohesive network of molecular defenders. What was most exciting, though, was the consistency. Despite differences in geography, species, temperature treatments, and tissue types, the same core genetic response kept showing up. This suggests a conserved heat-response toolkit—a genetic response shaped by years of evolution. The main “tool” (i.e. genes/pathways) identified in this core toolkit included:
- Heat Shock Proteins (HSPs): crucial for protein folding and protection against thermal damage.
- Ubiquitin–Proteasome System (UPS): involved in protein degradation and turnover.
- Alternative Splicing Mechanisms: allowing for versatile gene expression in response to stress.
Implications and Future Directions
Identifying this core genetic response provides a foundation for developing strategies to enhance the resilience of abalone. Potential applications include:
- Selective Breeding: utilizing genetic markers to breed heat-tolerant abalone strains.
- Early Stress Exposure: conditioning juveniles to moderate heat stress to build resilience.
- Gene Editing: exploring targeted interventions to bolster stress-response pathways.
We uncovered a core set of genes, the abalone’s genetic armour, consistently activated under heat stress. These insights unlock pathways for conservation and climate-resilient aquaculture. As marine heatwaves intensify in frequency and severity, findings like these are crucial not only for the conservation of abalone but also for protecting other vulnerable marine species.
BMC Genomics: Second dive into marine genomics
Follow the Topic
-
BMC Genomics

This is an open access, peer-reviewed journal that considers articles on all aspects of genetics, genomics and proteomics.
Related Collections
With Collections, you can get published faster and increase your visibility.
Genomics of microbiomes
The study of microbiomes has emerged as a dynamic field at the intersection of genomics, ecology, and health sciences. Microbiomes encompass the diverse communities of microorganisms, including bacteria, viruses, and unicellular eukaryotes, residing in various environments, such as the human body, food, soil, and aquatic systems. Understanding the genomic makeup of these microbiomes is crucial for unraveling their complex interactions with hosts and the environment. As advances in sequencing technologies, including single molecule sequencing, metagenomics and single cell omics, continue to evolve, researchers are better equipped to explore the rich genetic diversity (including pangenomes and epigenomes) and functional capacities of microbiomes across different ecosystems.
Investigating the genomics of microbiomes is pivotal for addressing critical questions in ecology, health, disease, and environmental sustainability. For instance, recent breakthroughs in the field have illustrated how microbiomes influence human health, from their roles in metabolism and immune function to their impact on mental health. Furthermore, understanding the genomics of environmental microbiomes can provide insights into biogeochemical processes and ecological resilience. As we deepen our knowledge of these microbial communities and develop computational biology methods to model their functionality, we stand to enhance our ability to harness their potential for applications in medicine, agriculture, and environmental management.
Future research in this area holds the promise of transformative advances in our understanding of microbiomes. The integration of multi-omics approaches, combining genomics with transcriptomics, proteomics, and metabolomics, may lead to a holistic view of microbial community dynamics and their functional implications. Additionally, developments in artificial intelligence and machine learning could further accelerate discoveries, enabling the identification of novel microbial functions and their roles in health and disease. As we continue to explore these intricate relationships, we can anticipate innovative strategies for harnessing microbiomes for therapeutic and environmental applications.
Topics of interest include, but are not limited to:
•Genomics and epigenomics of host-microbe interactions
•X-omics studies in environmental and host-microbiomes
•Advances in genomics of unculturable microorganisms
•Genome-guided development of synthetic microbiomes and consortia
•Microbiomes and environmental resilience: a genomic perspective
•The human microbiome: genetic diversity and functional potential
•Microbial adaptation and evolution in changing environments
•The role of microbiomes in antibiotic resistance and pathogenesis
•Computational and AI-driven methods for microbiome genomics
•Microbiome applications in sustainable agriculture and environmental management
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 07, 2026
Cattle genomics
BMC Genomics invites researchers to contribute to our Collection on Cattle genomics focusing on understanding the genetic makeup of bovine species, which is essential for improving livestock breeding and health. Advances in genomic technologies, such as next-generation sequencing and RNA sequencing, have enabled researchers to reveal insights into traits such as growth, meat quality, milk production, disease resistance, reproductive fitness, and overall adaptability in bovine genomes. This Collection aims to highlight the latest research developments in cattle genomics, encompassing both genomic and transcriptomic studies that contribute to the understanding of bovine biology.
Recent breakthroughs in genomic selection and precision breeding techniques have already shown promise in increasing efficiency in cattle production. The use of CRISPR-Cas genome editing, for example, has allowed for precise modifications to the cattle genome, introducing beneficial genetic variations without the linkage drag associated with traditional breeding methods. Additionally, the integration of omics technologies is paving the way for a holistic understanding of cattle biology, allowing for more effective management and breeding strategies. Studying the rumen microbiome using genomics, transcriptomics, proteomics, and metabolomics has revealed how microbial communities contribute to feed efficiency and nutrient absorption. This comprehensive approach enables targeted nutritional strategies that improve cattle health and productivity while reducing environmental impact. Such integrative studies facilitate the selection of cattle with optimal microbiome compositions, leading to more sustainable and efficient cattle production systems.
As research in cattle genomics progresses, we can anticipate the development of more sophisticated genomic tools that will enable precise manipulation of genetic traits in bovine populations. This may lead to enhanced resilience against diseases, improved reproductive performance, and better adaptation to changing environmental conditions. Ultimately, continued innovation in this field holds the potential to reform cattle production systems, ensuring sustainable livestock farming for future generations.
- Genomic selection in cattle breeding
- Transcriptomic analysis of bovine traits
- Pathogenicity and disease resistance genomics
- Advances in RNA-Seq applications for cattle
- Omics approaches to cattle health and productivity
- Genetic mapping of economically important traits
- Gene editing
- Metagenomics of the bovine gut microbiome
- Epigenetic regulation of growth and reproduction
- Comparative genomics of cattle and other livestock species
This Collection supports and amplifies research related to SDG 2, Zero Hunger.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer-review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: May 26, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in