Uncovering the hidden RNA targeting potential of CRISPR-Cas12a
Published in Healthcare & Nursing, Cancer, and Microbiology

Over the last decade, molecular diagnostics have undergone a transformative revolution with the help of scientists who have harnessed the immunological defenses of prokaryotes to detect human disease. At the forefront of this revolution lies a relatively obscure enzyme known as Cas12a.
Originally discovered as a part of the prokaryotic immune system called CRISPR1, Cas12a has soon emerged as a beacon of hope in the realm of medical diagnostics2. This is thanks to a remarkable property that it possesses known as ‘trans-cleavage’.
Trans-cleavage is an activity that allows Cas12a to not only target and cut specific DNA sequences of interest but also to indiscriminately cleave other unrelated single-stranded DNA (ssDNA) molecules in the vicinity, once activated. This unique property has opened up new avenues in the world of molecular diagnostics.
Here's how it works: When Cas12a, in conjunction with a small piece of RNA called the guide RNA, finds and binds to its target DNA sequence (usually the DNA of a disease-causing microbe), it undergoes a conformational change. This change activates its non-specific single-stranded DNA (ssDNA) cleavage ability. Thus, if a synthetic ssDNA reporter molecule (often tagged with a quenched fluorophore) is introduced into the mix, Cas12a will start cutting it up once it's activated by the target DNA. This cleavage of the reporter molecule can then be detected, signaling the presence of the target DNA sequence in the sample. In the last few years, the trans-cleavage property of Cas12a has been utilized to create point-of-care devices to detect a plethora of diseases such as COVID-19, HCV, malaria, and HIV, among others3-5.
Cas12a is primarily a DNA-cleaving enzyme. Even within the prokaryotic organisms that harbor it, it is speculated to confer immunity against DNA phages. Therefore, Cas12a-based diagnostic tests have been limited to detecting DNA targets but not RNA. Detecting RNA targets with Cas12a requires additional steps such as reverse transcription or strand displacement6-8, which can be expensive and add to the cost and complexity of the diagnostic test.
In this work, we have serendipitously uncovered a new and interesting RNA-targeting capability of Cas12a that has been hitherto unknown. Our story began when a graduate student in the lab (Swapnil Anekar), started speculating whether it would be possible to detect two different targets with Cas12a using the same guide RNA. Typically, the target binding region of the guide RNA for Cas12a is about 20-nt in length, of which the first 8-10-nt near the 5’-end is known as the PAM-proximal (Pp) region while the area near the 3’-end is known as the PAM-distal (Pd) region. Generally the guide RNA is designed such that the entire 20-nt guide binding region target the same molecule. The student wondered what would happen if we supplied a short 10-nt target to bind to the Pp-end and a separate 10-nt target to bind to the Pd-end - would the trans-cleavage activity still be triggered? Previous works have shown that a crRNA-DNA hybrid of at least 17-nt is required for stable Cas12a binding and cleavage9-11. Therefore, it was no surprise that individually these short targets were incapable of initiating the trans-cleavage. Interestingly, however, we observed when both targets were simultaneously supplied in a split-activator fashion, the trans-cleavage activity was indeed turned on.
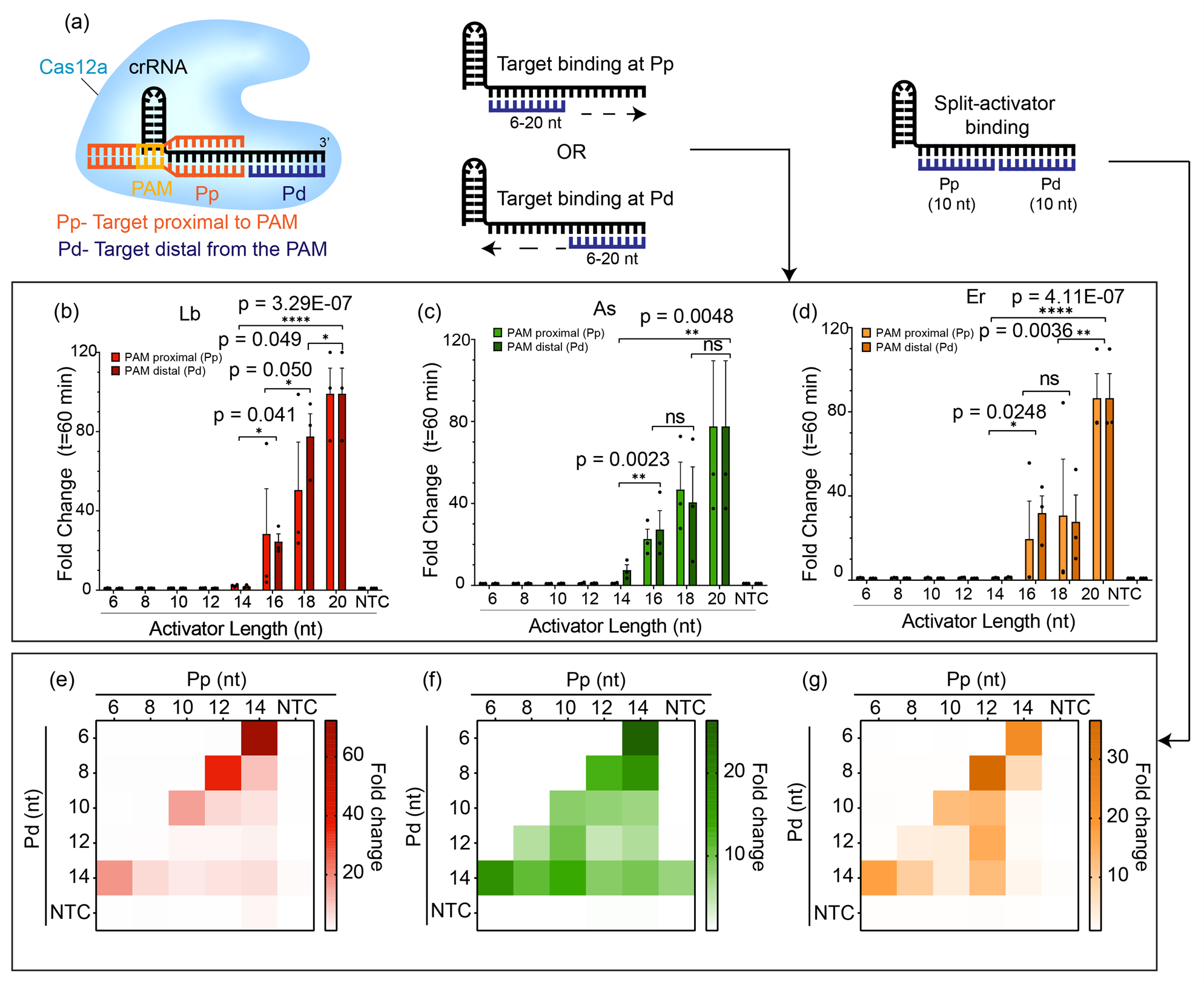
Fig.1: Short DNA activators fail to trigger trans-cleavage activity of Cas12a individually but are able to do so if supplied simultaneously as split-activators.
Next, we wondered what would happen if we switched one of the split activators to RNA instead of DNA. We expected the trans-cleavage to not be activated by the RNA substrates since Cas12a does not naturally tolerate RNA. However, we were surprised to see that contrary to our expectations, 6 different Cas12a orthologs were able to turn on trans-cleavage activity even with an RNA substrate binding to the Pd end of the guide RNA provided that a DNA substrate is simultaneously bound at the Pp. Remarkably, It seemed that with most Cas12 enzymes the Pp-end had a strict DNA-only preference, but the Pd-end could tolerate both RNA as well as DNA. The exception here was AsCas12a12, an ortholog of Cas12a derived from the bacteria Acidaminococcus sp. BVL36, which we observed was the only ortholog in our experiments that was capable of tolerating RNA at both the Pp-end as well as the Pd-end.
We developed these discoveries into a method of detection we reference as Split Activator for Highly Accessible RNA Analysis (SAHARA). When supplied with only a short synthetic DNA sequence complementary to the “seed” region, SAHARA detects picomolar concentrations of target RNA with no sample amplification, reverse transcription, or strand displacement. While some Cas proteins, such as Cas13, can detect solely RNA, they are not able to detect DNA. The ability to detect both DNA and RNA substrates in cis, to activate trans-cleavage is unheard of for any naturally occurring Cas proteins, and it enables a cheaper and more all-encompassing option for the detection of clinically relevant RNA targets.
To apply our discovery to more clinically important targets, we designed multiple guide RNAs targeting HCV RNA and miR-155. Hepatitis C virus (HCV) is an RNA-based virus that infects more than 170 million people worldwide13 causing liver inflammation and swelling. The microRNA-155 (or miR-155) is known to be overexpressed in breast cancer tissues14,15 and thus is used as an effective biomarker for early diagnosis. Early detection of both diseases is crucial for patients, as each undiagnosed day allows the diseases to further advance.
SAHARA provides a potentially inexpensive and easy option for the detection of these RNA targets. Our data suggested that the trans-cleavage was active with every guide RNA designed for HCV as well as miR-155, yet we observed varying levels of activity with different guides. This led us to analyze the guide designs and their corresponding targets further. We realized that when the target had increased secondary structure, the activity of Cas12a was reduced. This activity reduction is likely due to the complex secondary structure causing the target to become inaccessible to bind to the guide RNA. To improve the activity, we looked at previous studies16 that have shown that detection activity can be heightened for the Cas13a enzyme by pooling together multiple guide RNA. When following a parallel approach, SAHARA also saw greater activity with pooled guides for both miRNA-155 and HCV targets.
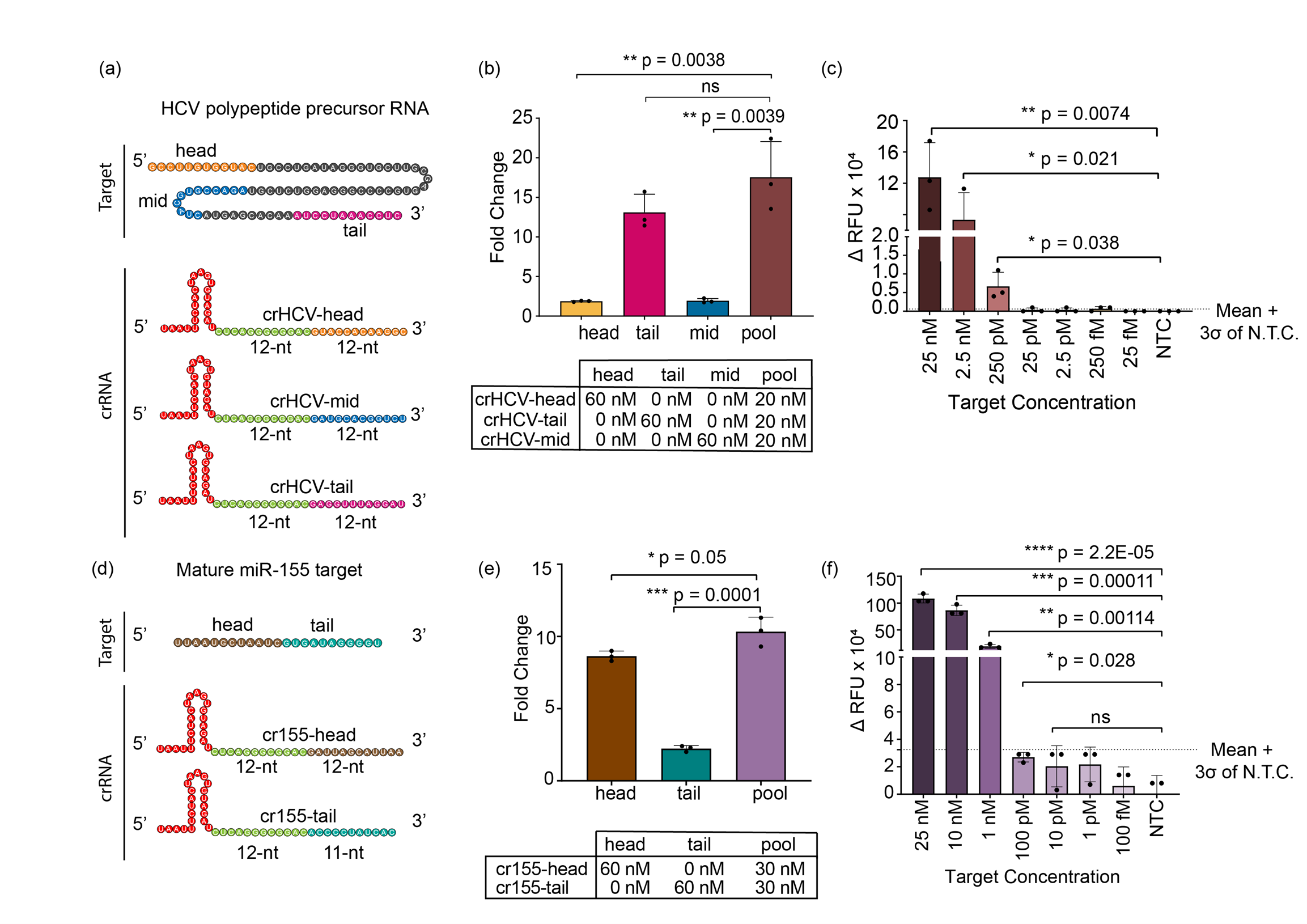
Next, we investigated the minimum concentration of S12 (the DNA activator that is needed at the Pp-end) for RNA detection with SAHARA. Observations showed that trans-cleavage activity correlated with S12 concentrations, suggesting its potential as an activity switch. Our data indicated that S12 DNA can selectively activate specific guide RNAs from a pool of different guides, enabling simultaneous detection of multiple DNA and RNA targets. By integrating SAHARA with Cas13b17 and using distinct fluorescent dyes such as FAM and HEX, we demonstrated SAHARA's capacity for multiplexed RNA detection of a plethora of mixed targets.
Although SAHARA still has some limitations that prevent it from being the industry standard for robust clinical detection, each discovery provides an incremental advance to the growing body of knowledge. The relative affordability, speed, and ease of CRISPR-based diagnostics have the potential to redefine the boundaries of healthcare; especially now that there is a novel route for more efficient RNA detection with SAHARA. With continued research into SAHARA, we hope to improve its sensitivity, specificity, and detection capabilities to increase its potential benefit in both clinical and point-of-care settings.
You can read more about our paper here: https://doi.org/10.1038/s41467-023-41006-1
References:
- Sternberg, S. H. & Doudna, J. A. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol. Cell 58, 568–574 (2015).
- Kaminski, M. M., Abudayyeh, O. O., Gootenberg, J. S., Zhang, F. & Collins, J. J. CRISPR-based diagnostics. Nat. Biomed. Eng. 5, 643–656 (2021).
- Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O. & Zhang, F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012 (2019).
- Zhang, W. S. et al. Reverse Transcription Recombinase Polymerase Amplification Coupled with CRISPR-Cas12a for Facile and Highly Sensitive Colorimetric SARS-CoV-2 Detection. Anal. Chem. 93, 4126–4133 (2021).
- Nguyen, L. T., Smith, B. M. & Jain, P. K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 11, 4906 (2020).
- Feng, W. et al. Integrating Reverse Transcription Recombinase Polymerase Amplification with CRISPR Technology for the One-Tube Assay of RNA. Anal. Chem. 93, 12808–12816 (2021).
- Zhang, W. S. et al. Reverse Transcription Recombinase Polymerase Amplification Coupled with CRISPR-Cas12a for Facile and Highly Sensitive Colorimetric SARS-CoV-2 Detection. Anal. Chem. 93, 4126–4133 (2021).
- Wang, X. et al. A sensitive and facile microRNA detection based on CRISPR-Cas12a coupled with strand displacement amplification. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 279, 121476 (2022).
- Jeon, Y. et al. Direct observation of DNA target searching and cleavage by CRISPRCas12a. Nat. Commun. 9, 2777 (2018).
- Stella, S. et al. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity. Cell 175, 1856-1871.e21 (2018).
- Singh, D. et al. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. 115, 5444–5449 (2018).
- Kleinstiver, B. P. et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016).
- Corey, K. E., Mendez-Navarro, J., Gorospe, E. C., Zheng, H. & Chung, R. T. Early treatment improves outcomes in acute hepatitis C virus infection: a meta-analysis. J. Viral Hepat. 17, 201–207 (2010).
- Mattiske, S., Suetani, R. J., Neilsen, P. M. & Callen, D. F. The Oncogenic Role of miR-155 in Breast Cancer. Cancer Epidemiol. Biomarkers Prev. 21, 1236–1243 (2012).
- Iorio, M. V. et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 65, 7065–7070 (2005).
- Fozouni, P. et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184, 323-333.e9 (2021).
- Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in