Unlocking the evolutionary mechanism of Intermolecular Diels-Alderases from Mulberry Trees
In organic synthesis, the Diels-Alder reaction is pivotal. Seeking eco-friendly alternatives, researchers explore natural Diels-Alderases, enzymes that catalyze this reaction. Xiaoguang Lei's lab uncovers evolutionary mechanisms in plant-derived Diels-Alderases, especially from mulberry trees.
Published in Chemistry, Ecology & Evolution, and Cell & Molecular Biology
paper: The evolutionary origin of naturally occurring intermolecular Diels-Alderases from Morus alba
The Diels-Alder reaction, esteemed as a luminous gem within organic synthesis, occupies an unparalleled niche in the synthesis of natural products. Its extensive applicability and deep-seated influence have propelled the search for naturally occurring enzymes capable of facilitating the Diels-Alder reaction, known as Diels-Alderases, into a pivotal area of investigation in biosynthesis. In today's biosynthesis sphere, researchers are fervently engaged in the discovery of Diels-Alderases from natural sources. Their objective centers on realizing an efficient and eco-conscious synthesis of Diels-Alder adducts through enzymatic catalysis.
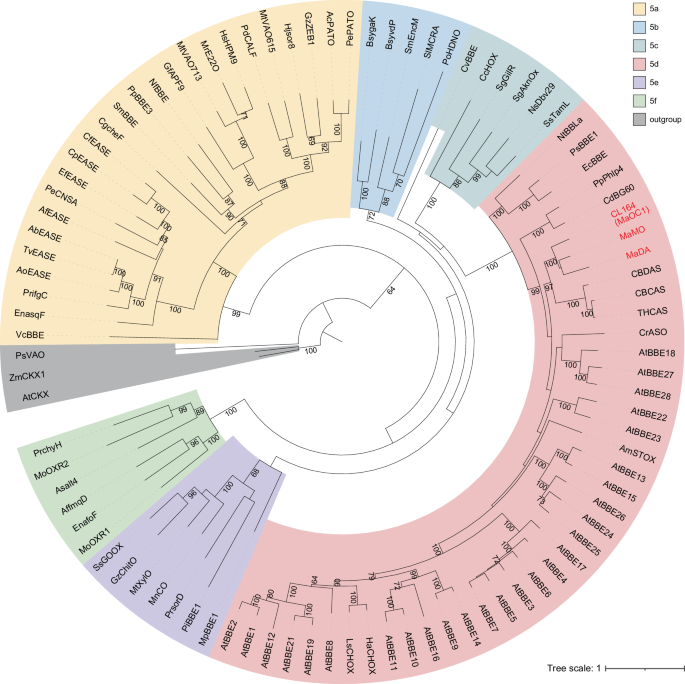
Within the spectrum of known Diels-Alderases, a notable lack of sequence and structural conservation among them poses a substantial challenge in the discovery of new Diels-Alderases. This issue is particularly pronounced in enzymes originating from plant sources, where the complexity of identification is significantly amplified. Consequently, a comprehensive investigation into the evolutionary mechanisms underpinning Diels-Alderases is imperative.
During the initial research phase in Xiaoguang Lei's laboratory, they identified the first plant-derived Diels-Alderases capable of catalyzing intermolecular Diels-Alder reactions, sourced from Morus alba. The team undertook exhaustive and methodical study of the catalytic mechanisms employed by these enzymes. Their significant discoveries have been disseminated through leading international publications, including Nature Chemistry (2020, 12, 620–628) and Nature Catalysis (2021, 4, 1059–1069). Building on these foundational investigations, the research group further discerned that the inter-molecular Diels-Alderase MaDA, originating from the Moraceae plant, shares considerable sequence and structural resemblances with the diene synthase MaMO. Both enzymes are classified under the BBE-like subfamily within the FAD-dependent oxidases. Notably, whereas the majority of enzymes in the BBE-like family predominantly engage in oxidation reactions, Diels-Alderases are exceptional for catalyzing non-oxidative processes. This unique trait suggests that MaDA may represent a novel functional adaptation within this enzyme family, a revelation that has garnered significant interest within the scientific community.
It's noteworthy that Diels-Alderases such as MaDA, Sol5, and PyrE3 are all hypothesized to have evolved from the FAD-dependent oxidases enzyme family, with MaDA firmly established as a pivotal member of this distinct group. A detailed study of the evolution of Diels-Alderase MaDA from FAD-dependent oxidases promises to unearth pivotal insights into the evolutionary origin and mechanisms underlying Diels-Alderases. In this context, the members of leilab have prioritized MaDA for their research endeavors.
To explore the evolutionary origins and mechanisms of Diels-Alderases more thoroughly, leilab implemented an array of technical methodologies, including ancestral sequence reconstruction, expression in insect systems, molecular dynamics simulations, and site-directed mutagenesis experiments. This multifaceted and systematic strategy unveiled the evolution from FAD-dependent oxidocyclases to Diels-Alderases and their functionally related diene synthases. The research team elucidated not only the specific mechanisms underpinning this evolutionary trajectory but also pinpointed critical residue mutations. Furthermore, they discovered nuanced adjustments in substrates that accompanied the enzyme's functional evolution, possibly facilitating a shift in enzymatic function. This pivotal research enhances our comprehension of the evolutionary development of Diels-Alderases and offers essential theoretical guidance for the discovery and engineering of novel Diels-Alderases.
Since the inaugural documentation of the Diels-Alder reaction in 1928, the quest to unearth biosynthetic enzymes integral to this reaction has engaged countless scientists across several decades. Yet, the discovery of Diels-Alderases has been remarkably limited. This profound investigation into the evolutionary mechanisms of Diels-Alderases sheds light on novel perspectives and strategies for the future mining and engineering of these enzymes. It heralds the promise of yielding an increased array of Diels-Alder-type adducts, thereby infusing new dynamism into the realms of biosynthesis and chemical synthesis.
The author states: Nature, replete with boundless energy, propels the evolution of life and molds countless marvels, weaving the vibrant fabric of our present reality. In response to the constantly fluctuating environment, plants synthesize structurally varied secondary metabolites, with enzymes serving as the architects of this biochemical diversity. Encoded by genes, enzymes perpetually evolve novel capabilities, thus fabricating structurally varied compounds for plants and contributing to the natural selection process where survival is reserved for the fittest. The evolution of enzymes not only generates a rich array of compounds but also drives the course of natural evolution and human advancement. Comprehending the intricacies of enzyme evolution enables us to collectively appreciate the magnificent spectacle of nature!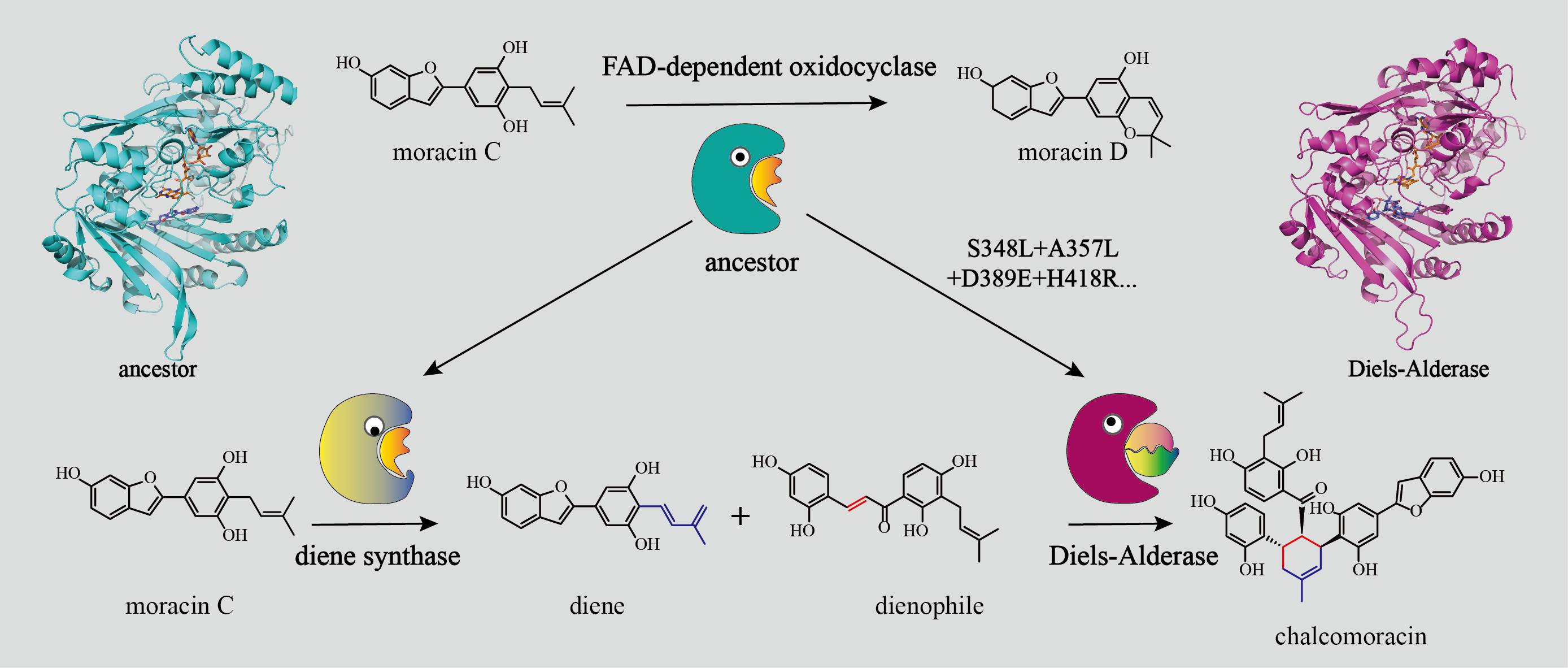

Follow the Topic
Enzymes
Life Sciences > Biological Sciences > Chemical Biology > Enzymology > Enzymes
Evolutionary Biology
Life Sciences > Biological Sciences > Evolutionary Biology
Biosynthesis
Physical Sciences > Chemistry > Biological Chemistry > Biosynthesis
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
A selection of recent articles that highlight issues relevant to the treatment of neurological and psychiatric disorders in women.
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
This Collection aims to bring together research from various domains related to neurodegenerative conditions, encompassing novel insights into disease pathophysiology, diagnostics, therapeutic developments, and care strategies. We welcome the submission of all papers relevant to advances in neurodegenerative disease.
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Xiaoguang Lei, Ph.D.
Professor of Chemistry and Chemical Biology
College of Chemistry and Molecular Engineering
Peking University (PKU), Beijing 100871, China
Senior PI of Peking-Tsinghua Center for Life Sciences
Senior PI of Shen Zhen Bay Laboratory
Editor of Bioorganic & Medicinal Chemistry
Phone: 86-10-62760292
Email: xglei@pku.edu.cn
Group website: http://www.chem.pku.edu.cn/leigroup/