Unveiling Single-Cell Resolution Insights into Epichaperomes in Tissues: Introducing the PU-TCO Clickable Epichaperome Probe
Published in Cancer, Neuroscience, and Protocols & Methods
Epichaperomes, intricate protein structures anchored on key chaperones, emerge as pivotal players in the landscape of cancers and neurodegenerative disorders such as Alzheimer's and Parkinson's. Understanding these structures is paramount for deciphering disease mechanisms and devising effective treatments.
While radiolabeled probes and chemoproteomics [1-3] have yielded valuable insights into epichaperomes, they often lack the precision, cellular resolution, and versatility needed to address fundamental questions about these biomolecular structures at the single-cell level within disease-affected tissues and organs.
In response to this challenge, Bay et al. unveil the PU-TCO clickable epichaperome probe and PU-NTCO control probe [4]. Through meticulous synthesis and rigorous characterization, conducted both in cell cultures and brain tissue, they have engineered a probe capable of discerning epichaperomes from chaperones with precision and sensitivity.
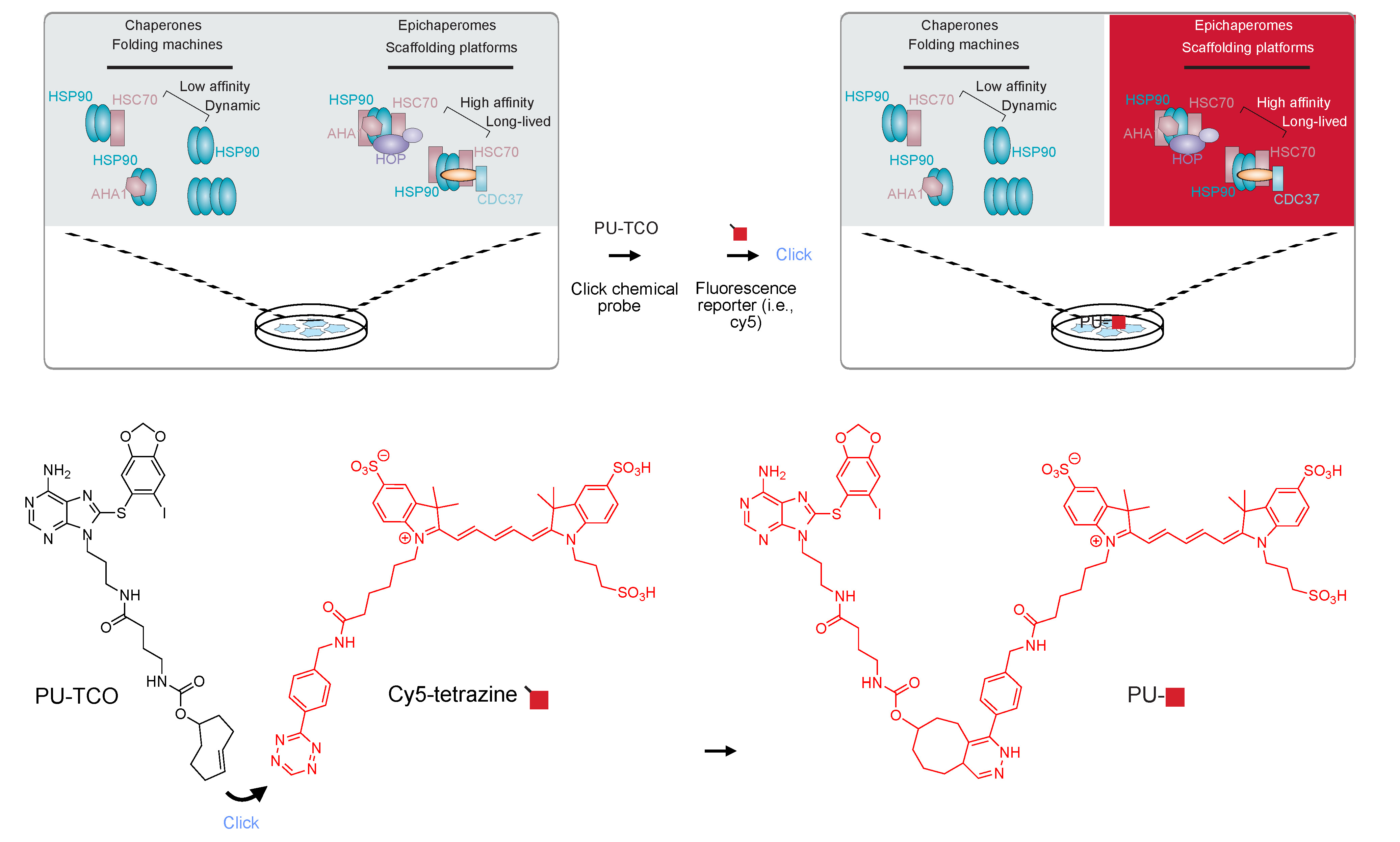
In a compelling proof-of-principle demonstration, Bay et al. have successfully deployed the PU-TCO probe in a mouse model of Parkinson's disease, capturing epichaperomes with cellular-level clarity. This breakthrough paves the way for exploring epichaperome biology amid the intricate web of biological systems, promising deeper insights into disorders.
In summary, the introduction of the PU-TCO clickable epichaperome probe marks a significant leap in disease research. This innovative probe promises precise imaging of epichaperomes in specific afflicted tissues and organs, leveraging the specificity of fluorescent probes and their compatibility with immunostaining techniques.
References
1. Pillarsetty N, Jhaveri K, Taldone T, Caldas-Lopes E, Punzalan B, Joshi S, Bolaender A, Uddin MM, Rodina A, Yan P, Ku A, Ku T, Shah SK, Lyashchenko S, Burnazi E, Wang T, Lecomte N, Janjigian Y, Younes A, Batlevi CW, Guzman ML, Roboz GJ, Koziorowski J, Zanzonico P, Alpaugh ML, Corben A, Modi S, Norton L, Larson SM, Lewis JS, Chiosis G, Gerecitano JF, Dunphy MPS. Paradigms for Precision Medicine in Epichaperome Cancer Therapy. Cancer Cell. 2019 Nov 11;36(5):559-573.e7. doi: 10.1016/j.ccell.2019.09.007. Epub 2019 Oct 24. PMID: 31668946; PMCID: PMC6996250.
2. Rodina A, Xu C, Digwal CS, Joshi S, Patel Y, Santhaseela AR, Bay S, Merugu S, Alam A, Yan P, Yang C, Roychowdhury T, Panchal P, Shrestha L, Kang Y, Sharma S, Almodovar J, Corben A, Alpaugh ML, Modi S, Guzman ML, Fei T, Taldone T, Ginsberg SD, Erdjument-Bromage H, Neubert TA, Manova-Todorova K, Tsou MB, Young JC, Wang T, Chiosis G. Systems-level analyses of protein-protein interaction network dysfunctions via epichaperomics identify cancer-specific mechanisms of stress adaptation. Nat Commun. 2023 Jun 23;14(1):3742. doi: 10.1038/s41467-023-39241-7. PMID: 37353488; PMCID: PMC10290137. https://www.nature.com/articles/s41467-023-39241-7
3. Inda MC, Joshi S, Wang T, Bolaender A, Gandu S, Koren Iii J, Che AY, Taldone T, Yan P, Sun W, Uddin M, Panchal P, Riolo M, Shah S, Barlas A, Xu K, Chan LYL, Gruzinova A, Kishinevsky S, Studer L, Fossati V, Noggle SA, White JR, de Stanchina E, Sequeira S, Anthoney KH, Steele JW, Manova-Todorova K, Patil S, Dunphy MP, Pillarsetty N, Pereira AC, Erdjument-Bromage H, Neubert TA, Rodina A, Ginsberg SD, De Marco Garcia N, Luo W, Chiosis G. The epichaperome is a mediator of toxic hippocampal stress and leads to protein connectivity-based dysfunction. Nat Commun. 2020 Jan 16;11(1):319. doi: 10.1038/s41467-019-14082-5. PMID: 31949159; PMCID: PMC6965647. https://www.nature.com/articles/s41467-019-14082-5
4. Bay, S.; Digwal, C.S.; Rodilla Martín, A.M.; Sharma, S.; Stanisavljevic, A.; Rodina, A.; Attaran, A.; Roychowdhury, T.; Parikh, K.; Toth, E.; et al. Synthesis and Characterization of Click Chemical Probes for Single-Cell Resolution Detection of Epichaperomes in Neurodegenerative Disorders. Biomedicines 2024, 12, 1252. https://doi.org/10.3390/biomedicines12061252



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in