A connectivity signature for glioblastoma – towards a molecular and phenotypical understanding of the deadliest brain tumor in adults
Published in Cancer and Genetics & Genomics
Glioblastoma (GB) remains an incurable brain tumor characterized by a median survival of 15-20 months and frequent therapy resistance 1. We have recently discovered that half of the tumor cells in GB interconnect into a cellular network through long membrane protrusions called tumor microtubes (TM)2–5. TMs facilitate communication among tumor cells, enable self-repair and proliferation, and ultimately convey resistance to chemotherapy, radiotherapy, and surgical interventions 4,5. However, determining and quantifying the extent of TM-connectivity in tumors is challenging, and the molecular understanding of the TM-network is limited. In our article recently published in Nature Communications, we aimed to address these aspects and introduced a clinically applicable gene expression signature for TM-connectivity in GB.
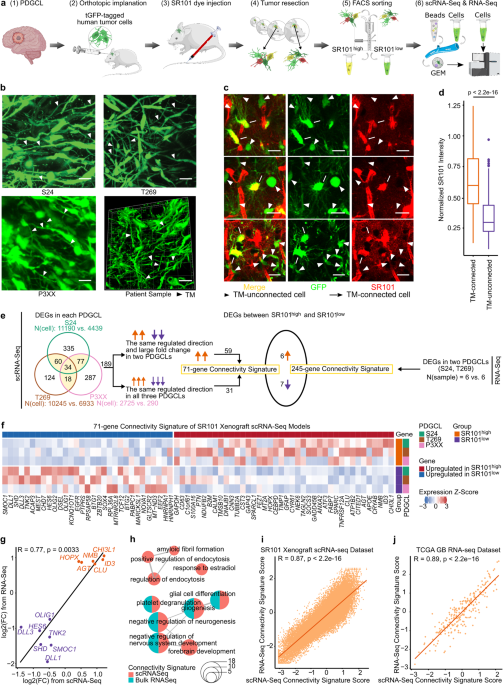
To characterize highly and lowly TM-connected GB cells on a molecular level we needed to identify and isolate cells of these respective populations. We achieved this by utilizing Sulforhodamine 101 (SR101), a fluorescent dye that preferentially labels TM-connected cells (Figure 1a-b). SR101high and SR101low cells from three xenografted human patient-derived glioblastoma cell lines (PDGCLs) were then separated by flow cytometry and subjected to both bulk and single-cell RNA-Seq (scRNA-Seq).
To develop a connectivity-related gene signature, we conducted differential expression analysis between SR101high and SR101low cells, independently for each PDGCL in the SR101 scRNA-Seq dataset. We identified a 71-gene signature, which we termed the “connectivity signature” (Figure 1c). To quantify the expression level of the signature, we developed a score that evaluated the average expression of the genes comprising the signature. This score effectively distinguished SR101high and SR101low cells in both bulk and single-cell RNA-Seq datasets (Figure 1d-e), and we named it “connectivity signature score”. In comparison to the scRNA-Seq-derived connectivity signature, we obtained a 245-gene signature from the SR101 bulk RNA-Seq dataset. Both signatures showed a common enrichment in neurogenesis-related gene ontology (GO) terms. Furthermore, the scores derived from both signatures exhibited a high correlation, not only in the SR101 scRNA-Seq dataset but also in the The Cancer Genome Atlas (TCGA) GB cohort RNA-Seq dataset.
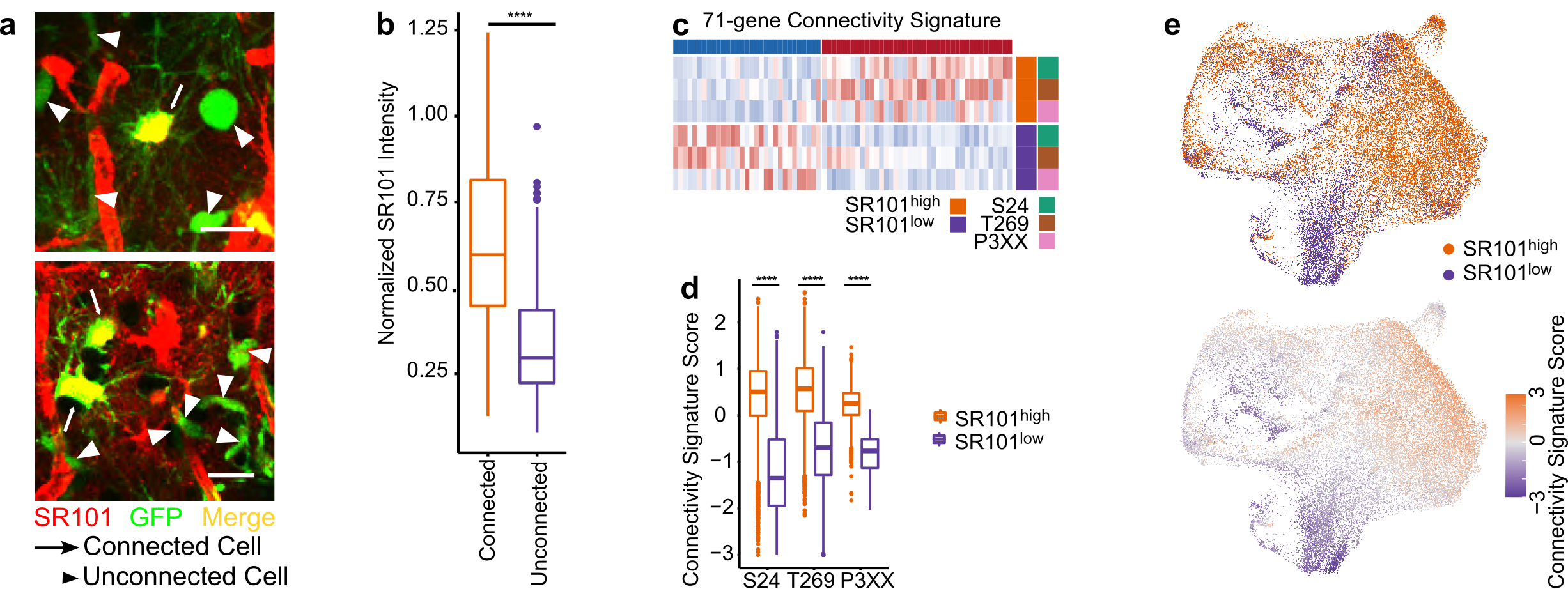
Figure 1. The connectivity signature is derived from a dye-transfer in vivo model. a. Two-photon microscopy images of SR101 intensity in TM-connected and TM-unconnected cells. b. Higher SR101 intensity in TM-connected cells compared to TM-unconnected cells. c. The 71-gene connectivity signature. d. The connectivity signature scores well-distinguished the SR101high and SR101low cells. e. Cells with higher connectivity signature score co-localized with the area of cells with SR101high intensity in UMAPs.
Empowered by scRNA-Seq technique, four distinct cellular states of GB cells have been described, with each state resembling the molecular profile of a specific healthy brain cell type 6. We thus delved deeper into each individual cell´s cell state in our SR101 scRNA-Seq dataset. We observed that the SR101high subpopulation predominantly comprised astrocytic-like cells (AC) and mesenchymal-like cells (MES), whereas the SR101low subpopulation exhibited a higher proportion of neuronal progenitor-like cells (NPC) and oligodendrocyte progenitor-like cells (OPC) (Figure 2a). Notably, we found that approximately half of the connectivity signature genes were also cell-state-defining markers, and that the connectivity signature score was higher in AC and MES compared to NPC and OPC cells (Figure 2b). This demonstrates a close relationship between connectivity and cell state. We corroborated this finding in two scRNA-Seq datasets of GB patients: Our newly generated 21-sample GB patient tumor scRNA-Seq dataset and a publicly available 110-sample GB patient tumor scRNA-Seq dataset (GBmap) 7.
To assess the relationship between connectivity and patient survival, we conducted survival analyses in the TCGA, Chinese Glioma Genome Atlas (CGGA), and The Glioma Longitudinal Analysis (GLASS) GB patients datasets. Our findings indicated that GB patients with higher connectivity signature scores experienced shorter overall survival times in all three cohorts (Figure 2c). Concluding from this, the connectivity signature score could serve as a prognostic marker for GB.
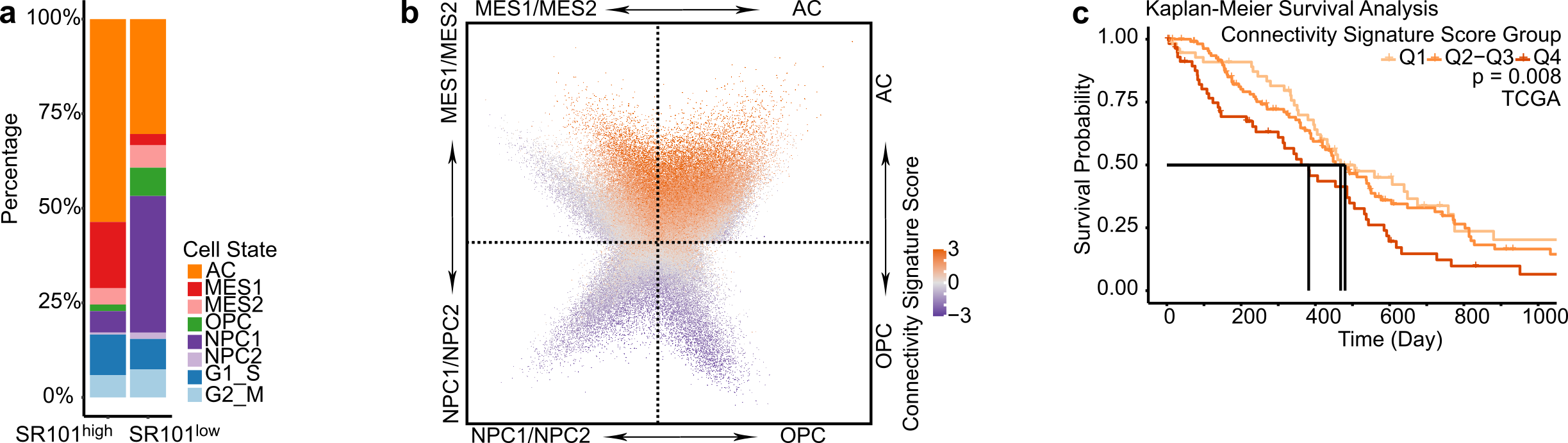
Figure 2. High connectivity associates with mesenchymal and astrocytic molecular profiles and impairs survival in GB patients. a. Different cell state composition between SR101high and SR101low samples. b. Higher connectivity signature score in cells with higher AC and MES signature scores. c. Higher connectivity signature score associated with shorter patient overall survival.
To validate the findings derived from the SR101 methodology, we conducted several orthogonal methodologies to investigate the interrelationship between phenotypical and molecular TM-connectivity. One of these experiments revealed that cells exhibiting a higher number of TM-connections also displayed more calcium activity and increased connectivity signature scores. Pharmacological interference with calcium activity, in contrast, was accompanied by a reduction in TM number and connectivity signature scores. When we induced TMs through modulation of certain cell culture parameters, we noted an increase in connectivity signature scores. Ultimately, the positive correlation of TM length and connectivity signature scores was corroborated in histological sections of FFPE GB patient tumor samples. Collectively, our findings indicated that cells with higher TM lengths and higher calcium activity exhibited higher connectivity signature scores and that the connectivity signature score serves as a reliable and robust indicator for the extent of TM-connectivity in samples.
The connectivity signature did not only include previously identified TM markers such as GAP43 and APOE but also unveiled novel candidate markers for TM-connectivity. One of them was CHI3L1. It exhibited higher expression in SR101high cells compared to SR101low cells in both SR101 bulk and single-cell RNA-Seq datasets. CHI3L1 further demonstrated the highest correlation with the connectivity signature score among the genes comprising the connectivity signature. Ultimately, CHI3L1 showed higher expression in samples with more calcium transients and exhibited higher histological staining intensity in samples with higher TM length.
To investigate if CHI3L1 is not only a marker but also a driver of TM-connectivity, we applied a CHI3L1 blocking antibody to PDGCLs, which resulted in reduced TM lengths. Additionally, overexpressing CHI3L1 in PDGCLs led to longer TMs and increased the number of TM connections (Figure 3a-b). Diving into the mechanism of CHI3L1-driven TM-connectivity, the molecular profiles of CHI3L1 overexpression cells were assessed by RNA-Seq, proteomics, and phosphoproteomics. The results showed that overexpression of CHI3L1 led to an increased AC signature score, decreased NPC1 signature score, and a higher connectivity signature score at both RNA and protein levels (Figure 3c). Interestingly, overexpression of CHI3L1 also led to higher phosphorylation in a previously identified TM driver, GAP43 (Figure 3d). These findings support the role of CHI3L1 as a connectivity driver.
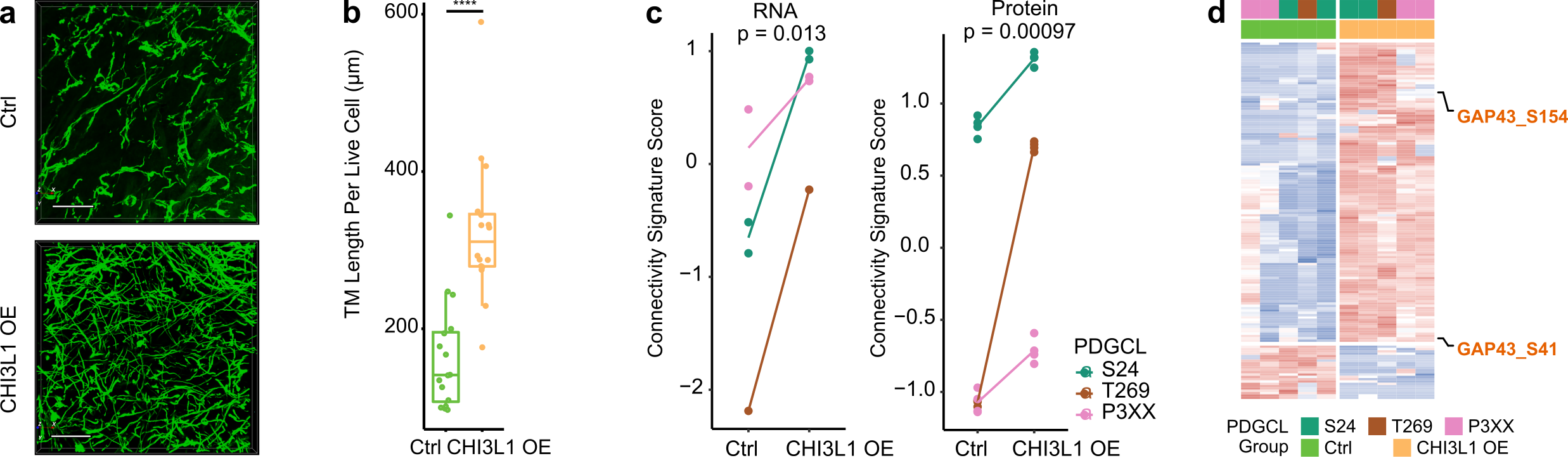
Figure 3. CHI3L1 overexpression is accompanied by increased phenotypical and molecular connectivity. a. 3D micrographs showed longer TM lengths and more TM connections in CHI3L1 overexpressed samples. b. Longer TM lengths in CHI3L1 overexpressing samples. c. Higher connectivity signature score in CHI3L1 overexpression cells at both RNA and protein levels. d. CHI3L1 overexpression cells had higher GAP43 phosphorylation.
In summary, we introduced and validated a clinically applicable connectivity signature for GB. Our results revealed that AC and MES populations are characterized by higher connectivity signature scores, and patients with higher scores have a dismal survival. Additionally, we identified CHI3L1 as a novel marker and driver for TM-connectivity.
References
- Alexander, B. M. & Cloughesy, T. F. Adult Glioblastoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 35, 2402–2409; 10.1200/JCO.2017.73.0119 (2017).
- Xie, R. et al. Tumor cell network integration in glioma represents a stemness feature. Neuro-oncology 23, 757–769; 10.1093/neuonc/noaa275 (2021).
- Venkataramani, V. et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573, 532–538; 10.1038/s41586-019-1564-x (2019).
- Weil, S. et al. Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro-oncology 19, 1316–1326; 10.1093/neuonc/nox070 (2017).
- Osswald, M. et al. Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98; 10.1038/nature16071 (2015).
- Neftel, C. et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835-849.e21; 10.1016/j.cell.2019.06.024 (2019).
- Ruiz-Moreno, C. et al. Harmonized single-cell landscape, intercellular crosstalk and tumor architecture of glioblastoma. Biorxiv. 10.1101/2022.08.27.505439 (2022).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in