Algal blooms from many angles
Published in Earth & Environment

Algal blooms are hotspots of marine primary production and play central roles in microbial ecology and global nutrient cycling, but it is typically hard to predict exactly when and where a bloom will occur, and therefore, time-resolved studies of blooms are difficult to accomplish. In particular, the causes of bloom termination and their consequences on the marine ecosystem remain unresolved. This is where so-called mesocosm experiments are extremely powerful: by isolating a natural inoculum in impermeable bags (Figure 1), they enable repeated sampling of a complex community in a controlled manner. Here, our goal was to observe an algal bloom from the microscopic (cell counts, microbial community composition, etc.) to the macroscopic (ecosystem-level changes, biogeochemistry) scale to understand what happens when coccolithophore blooms get infected by viruses: how does viral infection alter the dynamics of the blooming algae1, the chemical environment in this transient miniature ecosystem2,3, and the surrounding microorganisms that depend on it, ultimately affecting the fate of carbon in the ocean?

Figure 1: Assaf Vardi and Guy Schleyer filling a mesocosm bag with 11,000L of natural Fjord water that will become our “natural inoculum”
In May 2018, our team, around 20 scientists from nearly ten labs from Israel, Germany, Spain, France, and the US, met in a small village near Bergen in Norway. In a nearby fjord, natural coccolithophore blooms of Emiliania huxleyi occur typically on a yearly basis, and the local Aquacosm facilities are equipped to assist researchers in studying algal blooms. Indeed, instead of waiting for a natural bloom to occur, we performed the mesocosm experiment: fill seven 11,000L bags with fjord water, add nutrients, and wait (and hope!) for a coccolithophore bloom to happen – all while taking careful measurements of various biological and chemical parameters twice a day. Our group consisted of many leading experts from various backgrounds in biology and chemistry, bringing together different techniques such as metabolomics, single cell sequencing, biogeochemistry, aerosols measurement, and glycobiology.
Waiting and hoping was indeed all we could do. With no control over the initial composition of microbial communities in our bag, we can only hope that enough E. huxleyi cells will start to divide to create a bloom. Thankfully, the first signals of calcified cells appeared in the flow-cytometer after ten days. But those few dots on the flow-cytometer plots were too few to be taken seriously; less than 10 events per gate cannot be trusted! Those ten first days felt very, very long, especially when you are in charge of reporting the daily counts to the whole consortium in the evening meetups – everyone is dreading the spectre of “what if we get no bloom”.
When we saw the first signs of coccolithophores, we were halfway through the mesocosm, and because our sampling regime was pretty intense, signs of fatigue inevitably appeared. Conducting the core morning sampling or common duties such as grocery shopping becomes harder, suggestions to improve the daily routines are formulated with less diplomacy than at the beginning – a very natural dynamic for such a field experiment that often gets solved easily around a beer and a few jokes. At last, the doubts dissipated, as the water in our bags turned milky, due to the calcium carbonate shells of the coccolithophores (Figure 2). But we did not have much time to contemplate, as the joy of seeing coccolithophores is quickly replaced by the adrenaline of hopefully getting viral infections.
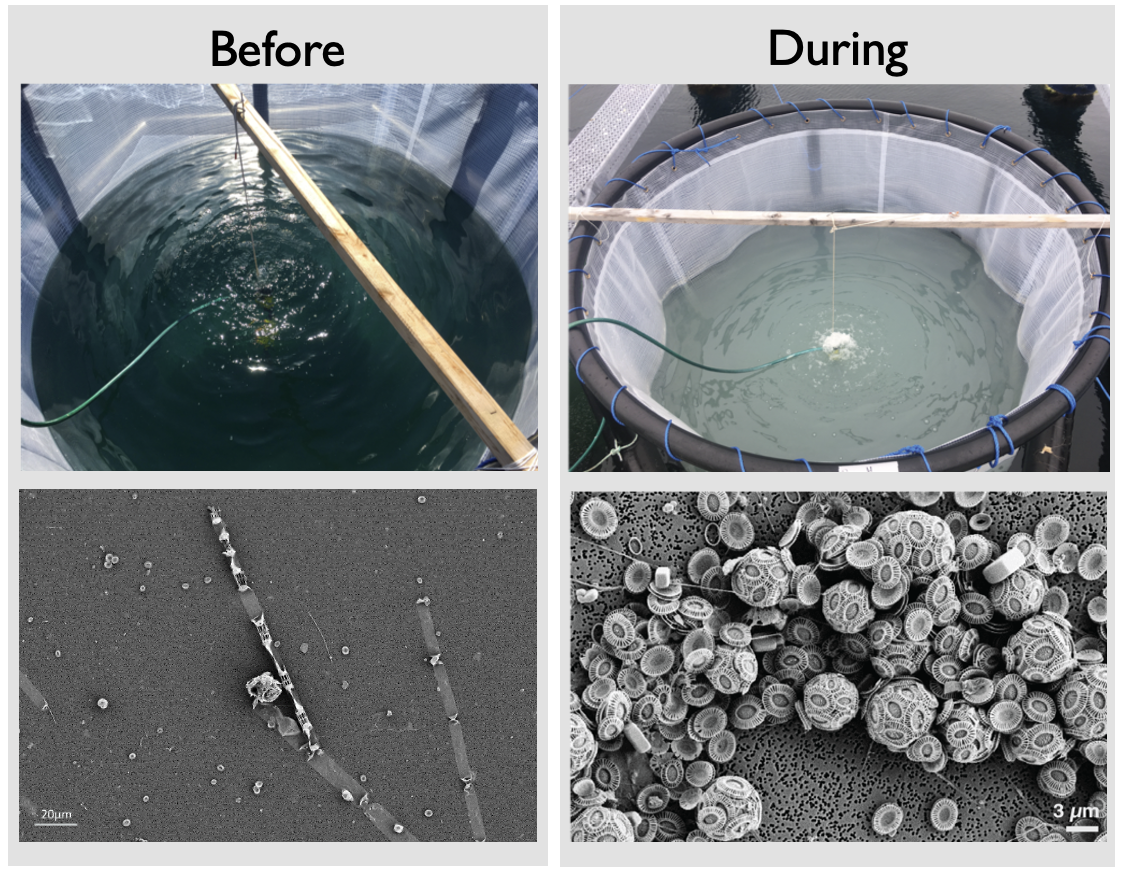
As the days passed, our coccolithophore concentrations reached ridiculously high numbers. First signs of giant viruses appeared in the cytometry gates, but we would have to wait for further analysis to be sure. After about 30 days in Norway, the team went back to their own labs to analyse the treasure trove of data we had collected, without forgetting to celebrate in style (Figure 3).

Figure 3: This picture was taken after the last sampling day, naturally.
More than a year later in October 2019, the whole team was hosted by the Vardi Lab at the Weizmann Institute to discuss initial findings. Each team member highlighted different parts of the experiments, from microbiome compositions at different size fractions to biogeochemical measurements, flow cytometry measurements, to intracellular measurements of active viral infection in single cells. Our task was now to combine all these different data streams and extract signals from this mass of data.

The on-site signals were quickly confirmed after combining the flow cytometry with sequencing data: two phytoplankton blooms had taken place, and the second bloom – the sought-after E. huxleyi bloom – had clearly taken different paths in different mesocosms. Although all mesocosms showed the beginnings of a bloom of E. huxleyi, the blooms were cut short in two bags. Through flow cytometry and qPCR, it was quickly determined that those bags also had very high abundances of virus infecting E. huxleyi. Success!
Analysing the composition of the surrounding microbiomes (both planktonic bacteria and picoeukaryotics) showed a beautiful succession of eukaryotes and bacteria at different rates and taxonomic scales, showing the highly dynamic nature of phytoplankton blooms: within a few days, the initial eukaryotic microbiome had almost completely been replaced by an entirely different community. Our highly resolved time-series sequencing gave us unprecedented insight into the microbiome dynamics during algal blooms.
Given that phytoplankton blooms release a lot of carbon that can be used by the surrounding bacteria, we expected a massive impact of E. huxleyi bloom and in particular its viral infection on the bacterioplankton, a key ecosystem process termed the “viral shunt”. However, a first surprise came from the flow cytometry data: whereas bacterial abundances increased more than 10-fold during the first bloom, they changed much more subtly during the E. huxleyi blooms, and despite the differences in the E. huxleyi dynamics between mesocosms, the microbiome compositions seemed to be relatively unaffected by the viral infection. One player stood out, however: Thraustochytrids, small protists that are known to degrade organic matter such as dead phytoplankton cells. These Thraustochytrids were increasing massively in abundance as the E. huxleyi bloom was starting to end, and even more so in the infected mesocosms. This could explain why bacteria hadn’t increased in abundance as much as expected during the E. huxleyi bloom: Thraustochytrids compete with bacteria for the organic carbon released by dying phytoplankton, and they seemed to be winning, rivaling bacteria by biomass towards the end of the experiment. Thus, we had identified Thraustochytrids as a wholly unappreciated, but essential heterotrophic player in the microbial ecosystems surrounding coccolithophore blooms.

Figure 5: During the bloom demise, E. huxleyi cells shed their coccoliths, covering the entire SEM filter !
Next, we focused on the biochemical measurements to assess the effects of viral infection on the biogeochemistry of algal blooms. When we looked at TEP, extracellular polysaccharides that are thought to play an important role in promoting aggregating of organic matter into particles, we noticed that their concentrations were lower in the infected bags relative to the others, but so were the abundances of E. huxleyi in those bags. How could we decide whether infected cells produced more or less TEP than uninfected cells? We developed a model that describes the amount of TEP over time based on the abundances of various phytoplankton which showed that the infected bags actually produced more TEP per cell! Integrating this model with data showing that only about one in five cells was infected1, we could estimate by how much: about 4 times as much TEP. This observation was crucial in interpreting our experiment in the context of the impact of viral infection on carbon export.
Thus, by combining various data streams, from single-cell measurements to characterisations of microbial community compositions to biogeochemical measurements, we were able to gain deeper insight into the biotic and abiotic dynamics and processes governing algal blooms. For this project, this combination of data streams–- made possible by the breadth of scientific expertise in our team–- turned out to be crucial to make sense of the data. This highlights the value of working in interdisciplinary teams, including microbiologists, oceanographers, chemical ecologists and bioinformaticians to discover patterns and gain insights that would remain hidden to each of them individually. Plus, it’s much more fun to do science in a team!
Flora Vincent, Group Leader at EMBL, Germany
Matti Gralka, Assistant Professor at VU-Amsterdam, Netherlands
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in