Antibiotics fuel the rise of vancomycin-resistant enterococci intestinal colonisation by remodelling the metabolic landscape of the gut microbiome
Published in Microbiology and Protocols & Methods
One of the great challenges of the 21st century will be managing essential antibiotic use to avoid the development of antibiotic-resistant pathogens. Since their clinical emergence, vancomycin-resistant enterococci (VRE) have become highly problematic opportunistic pathogens that cause difficult-to-treat and potentially fatal infections such as bloodstream infections, urinary tract infections, and endocarditis. Treating patients with broad-spectrum antibiotics significantly increases the risk of VRE colonising their intestines. Antibiotic-treated intestines act as a reservoir for VRE, enabling VRE to spread from the intestine to other sites in the body such as the bloodstream.
Antibiotic treatment is intended to treat infections, but how does antibiotic exposure increase a patient’s susceptibility to VRE intestinal colonisation and the subsequent development of VRE infections? Healthy individuals are protected from VRE intestinal colonisation because they have diverse mixtures of microorganisms that colonise their intestine (the “gut microbiota”) that protect them from VRE intestinal colonisation (termed “colonisation resistance”). However, broad-spectrum antibiotics not only kill pathogens, but they also kill protective members of the gut microbiota as well, promoting VRE intestinal colonisation.
Can a treatment or prophylactic be designed to prevent VRE intestinal colonisation without further disrupting essential functions of the gut microbiota? Our research aimed to determine which aspects of an antibiotic-treated intestine promote VRE intestinal colonisation. Using ex vivo human faecal cultures and mouse models, we examine the nutritional and metabolic environment that VRE experience in the antibiotic-treated intestine and how this promotes VRE growth. Finally, we identify potential targets for future microbiome therapeutics to combat VRE intestinal colonisation.
Antibiotics reduce competition for nutrients
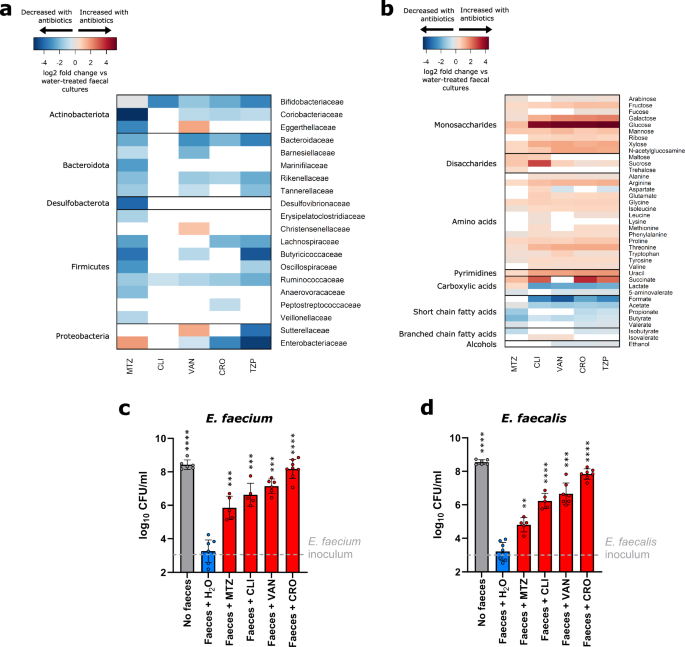
Healthy individuals are resistant to VRE intestinal colonisation because they have a gut microbiota that has high diversity and density. The competitive and collaborative nature of commensal gut microbial species ensures opportunistic pathogens fail to grow and colonise the intestine. Using ex vivo faecal cultures from healthy donors, we measured the impact of broad-spectrum antibiotics on the composition and metabolism of the gut microbiome. 16S rRNA gene sequencing and 16S qPCR data showed that antibiotic treatment led to a decrease in the abundance of many bacterial families, including striking decreases of key commensal bacterial families such as Ruminococcaceae and Bifidobacteriaceae.
Consequently, nutrients otherwise consumed by these commensal bacteria significantly increased in concentration, including monosaccharides, disaccharides, and amino acids. These nutrients could support the growth of VRE in an antibiotic-treated intestine.
VRE can use nutrients that were enriched in an antibiotic-treated intestine
The nutrient utilisation abilities of VRE strains were measured to determine how VRE can grow in a nutrient-enriched antibiotic-treated intestine. We tested nutrients that increased in concentration following antibiotic treatment to understand the carbon and nitrogen utilisation abilities of VRE strains. Both vancomycin-resistant Enterococcus faecalis and Enterococcus faecium used a variety of monosaccharides and disaccharides as carbon sources and used amino acids as nitrogen sources, with slight differences between the different species.
When grown in the presence of all nutrients identified to increase in faecal cultures following antibiotic treatment, VRE strains demonstrated a clear preference for certain nutrients, which were consumed before others. Glucose, mannose, and trehalose were rapidly depleted within the first several hours of growth. To maintain growth after favoured nutrients were depleted, VRE switched to use less preferred nutrients. This adaptability could be an essential tactic for colonisation within the gut environment.
Decreases in microbial metabolite production permits VRE intestinal colonisation
While nutrient availability increased following antibiotic treatment, microbial metabolites (such as short- and branched-chain fatty acids) decreased in concentration. These metabolites, produced by commensal microbiota members, can inhibit pathogen growth through direct antimicrobial mechanisms as well as indirectly by maintaining gut barrier function and modulating oxygen concentrations.
We evaluated the impact of 10 microbial metabolites at low, average, and high concentrations found in healthy individuals. While we saw some inhibition by acetate, propionate, butyrate, and valerate individually on VRE growth, the impact was incomplete. However, a mixture of these metabolites achieved complete or near-complete suppression of VRE strains.
VRE occupy overlapping but distinct intestinal niches
Previously published research by our lab on carbapenem-resistant Enterobacteriaceae (CRE) showed similar findings to our work with VRE. As such, we wanted to explore whether the overlapping nutrient-defined niches of these antibiotic-resistant pathogens resulted in competition or coexistence.
Using a one-to-one co-culture design, we showed that vancomycin-resistant E. faecalis and E. faecium grew to high levels alongside carbapenem-resistant Escherichia coli, Klebsiella pneumoniae, and Enterobacter hormaechei strains in media supplemented with nutrients enriched in antibiotic-treated faecal culture. Understanding this overlap provides crucial insights for designing therapeutics targeting multiple opportunistic pathogens simultaneously.
Future avenues for therapeutic design
The findings from this study have vital implications for developing microbiome therapeutics that harness the gut microbiome to suppress VRE growth. Strategies could aim to reintroduce key commensal species capable of outcompeting pathogens for essential nutrients and/or restore inhibitory metabolite concentrations. Our work suggests that short-chain fatty acids themselves could act as a beneficial therapeutic—either as a standalone treatment or combined with targeted gut commensal strains—to directly suppress VRE and other antibiotic-resistant pathogens.
While antibiotics have revolutionised medicine, they are not without serious and growing risk. Our research underscores the importance of understanding not just which bacteria are lost following antibiotic treatment, but what functions related to colonisation resistance are lost with them. By mapping both the microbial composition as well as the nutrient and metabolite landscape that VRE encounter following antibiotic treatment, we hope to empower the design of targeted microbiome therapeutics that restore gut microbiome functionality, prevent pathogen intestinal colonisation, and reduce the risk of devastating infections by antibiotic-resistant pathogens.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in