Deep learning-based microsatellite instability predictor from prostate cancer whole-slide images
Published in Cancer, Computational Sciences, and Genetics & Genomics
DNA repair pathways are a critical mechanism for cells to maintain the fidelity of their genome as they divide. Loss of DNA repair, which allows mutations to accumulate, can be an initiating or potentiating factor in cancer development and also, critically, a target for cancer immunotherapy. The DNA mismatch repair (MMR) genes, MLH1, PMS2, MSH2, and MSH6, form one such crucial pathway. Lynch syndrome, a heritable condition leading to increased risk of certain cancers, is caused by germline mutations in the MMR pathway. More commonly, two copies of an MMR gene are inactivated by somatic alterations. When deficiency of mismatch repair (dMMR) occurs, many mutations accumulate in the cell and a phenotype of high microsatellite instability (MSI-H) can be observed.
Next-generation sequencing (NGS)-based profiling or other molecular tests can be used to detect the MSI-H phenotype, while immunohistochemistry (IHC) can detect dMMR. While MSI/MMR testing is common for colon and endometrial cancers, due to a number of factors such as low pre-test probability, tissue availability, and cost considerations, testing is not routinely done for patients with other cancer diagnoses even though there is a tissue agnostic approval for immune checkpoint inhibitor therapy that can improve outcomes in this population. In prostate cancer, for instance, the MSI-H phenotype is uncommon and MSI-H testing is recommended only in metastatic castrate-resistant prostate cancer in guidelines from the National Comprehensive Cancer Network (NCCN). Nevertheless, up to ~2-3% of prostate cancer patients may harbor this important biomarker and these patients may be missing out on the opportunity for possible treatment options.
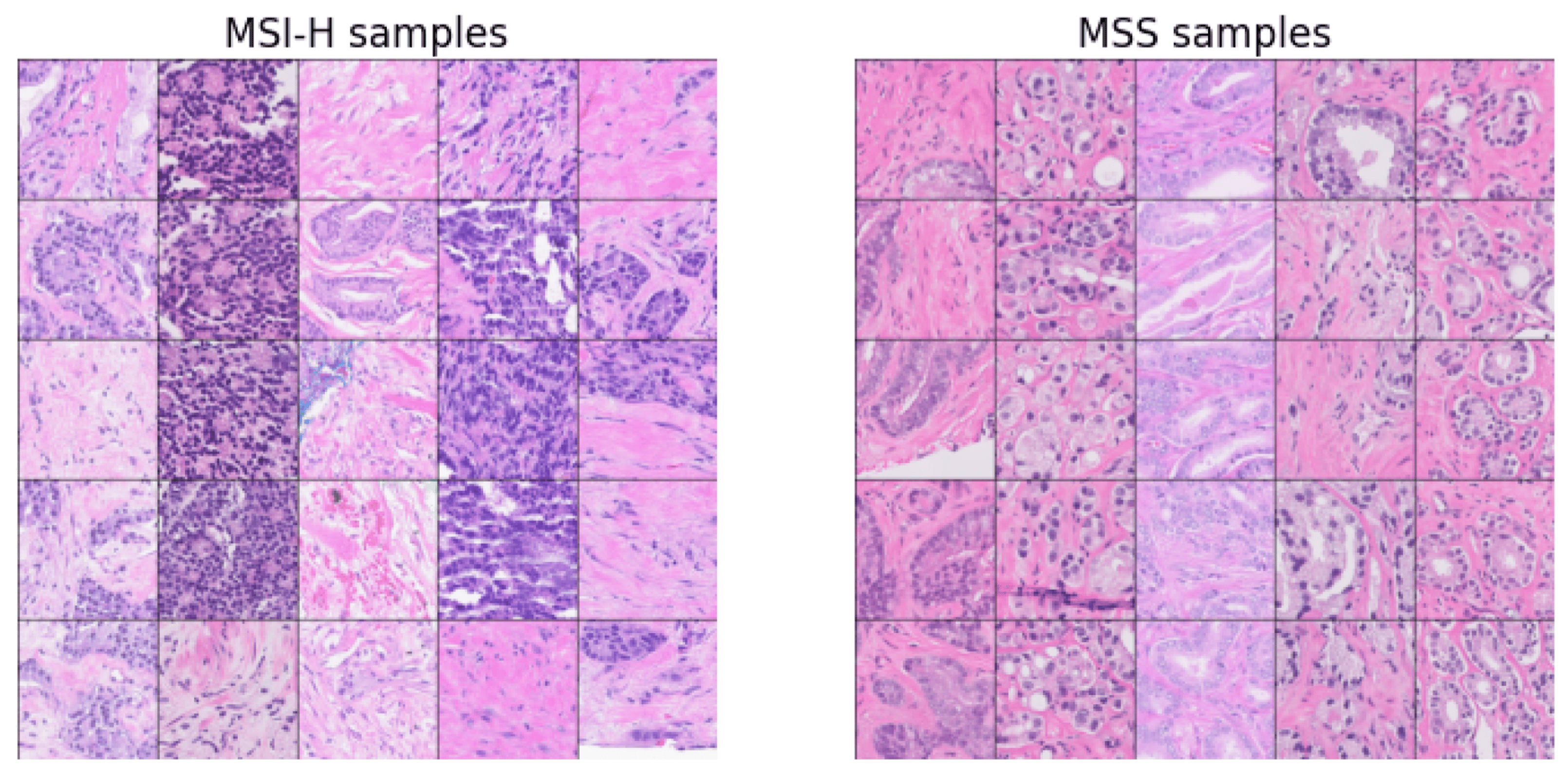
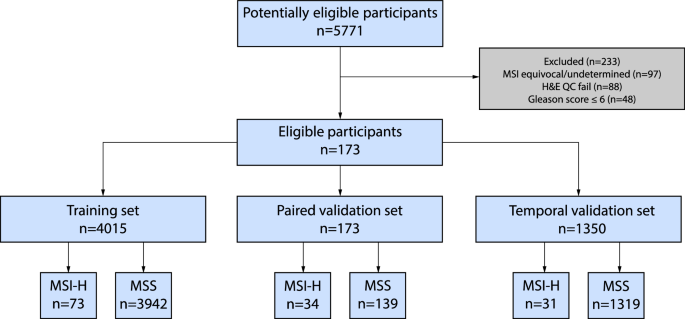
Cancer diagnosis nearly always involves a tissue biopsy with hematoxylin and eosin (H&E) stained slides, which are increasingly digitized as whole slide images (WSIs). We thus developed a digital pathology algorithm that analyzes WSIs to predict MSI-H status in prostate cancer patients with the intent to prioritize patients for confirmatory MSI/MMR testing. We trained an attention-based multiple instance learning model on a large real-world cohort of 4015 WSIs from primary prostate cancer biopsy or resection samples. For our “source of truth” used in model training and validation, we relied on MSI-H status calls from the Tempus xT DNA-seq assay. Our imaging-based algorithm analyzes features in many small image tiles from each WSI and aggregates them to generate a slide-level prediction score. The algorithm was evaluated on a real-world cohort of 1350 samples sequenced after the cohort used to develop the model and demonstrated good performance in predicting MSI-H, with an area under the receiver operating characteristic curve (a measure of classification accuracy) of 0.72. The algorithm also achieved similar performance on a paired dataset of internally and externally stained and scanned images, thereby showing effective generalizability to staining and scanning variations.
As part of our mission to provide precision medicine to patients with cancer, Tempus AI, Inc. has validated and deployed p-MSI, an updated version of the digital pathology algorithm reported in this publication, as part of the Tempus xT solid tumor DNA sequencing assay. The p-MSI algorithm is run for all prostate cancer patients who do not receive an MSI result but have sufficient tissue remaining to specifically assay MSI or dMMR status by other methods (MMR IHC only requires 100 cells/slide to interpret). The p-MSI algorithm identifies prostate cancer patients whose tumors are more likely than the average prostate cancer patient to be MSI-H. These patients could benefit from alternative confirmatory testing such as MMR IHC.
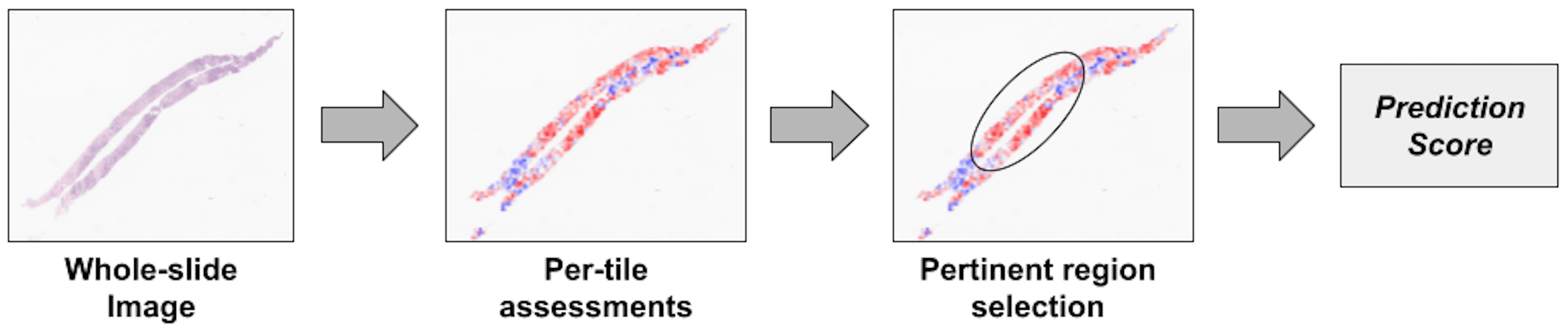
For this updated p-MSI assay, a Tempus pathologist selects pertinent regions for the model to consider, further reducing the influence of regions containing imaging artifacts or low tumor purity on the prediction. Subsequently, the algorithm analyzes tiles within these regions and generates a prediction score for the case. Following review and agreement by a pathologist, predicted MSI-H cases are communicated to clinicians. While similar to the work we published, the updated p-MSI algorithm demonstrates an area under the curve (AUC) of 0.82 in distinguishing MSI-H tumors from microsatellite stable (MSS) tumors in the analytical validation.
Tempus AI’s effort in developing digital pathology AI algorithms extends beyond predicting MSI-H in prostate cancer. Our researchers have been developing histogenomic models to infer a suite of other biomarkers for several cancer types from H&E stained slides. Molecular alterations detected by NGS have predictive, prognostic, and diagnostic value across various cancer types. Therefore, predicting molecular alterations from H&E WSIs is potentially impactful for identifying patients who likely have certain biomarkers and may benefit from confirmatory testing to evaluate their eligibility for targeted therapies and other downstream implications. Ultimately, digital pathology paired with artificial intelligence tools, which have advanced rapidly in recent years, may one day help predict the presence of many tumor molecular biomarkers, rapidly screen patients for molecular alterations, and prioritize patients for further testing.
Follow the Topic
-
npj Precision Oncology

An international, peer-reviewed journal committed to publishing cutting-edge scientific research in all aspects of precision oncology from basic science to translational applications to clinical medicine.
Related Collections
With Collections, you can get published faster and increase your visibility.
AI Approaches in Drug Design
Publishing Model: Open Access
Deadline: Mar 31, 2026
Genomic Instability
Publishing Model: Open Access
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in