µDeSCRiPTor: Revolutionizing tuberculosis therapeutics through single-cell biology
Published in Bioengineering & Biotechnology, Chemistry, and Microbiology

In the relentless battle against microbial pathogens, tuberculosis presents a formidable challenge1. This is due to the inherent ability of Mycobacterium tuberculosis to evade the host and therapy, and is exacerbated by the relentless spread of antibiotic resistance. In our efforts to combat this global health threat, we have embarked on a search for better therapeutic options. The fact that our efforts could also help combat other infectious diseases affected by antibiotic resistance gave us additional motivation. Our journey was driven by innovation, interdisciplinary collaboration, passion and commitment, and led us to develop a breakthrough strategy we call µDeSCRiPTor, which we recently reported on in the journal Nature Communications https://rdcu.be/dH9il.
A fundamental feature of any clonal bacterial population is the presence of phenotypic variation, which underlies the adaptive success of microorganisms2. As this is a multifactorial phenomenon, it is particularly complex to capture and interpret, and it needs to be studied at the single-cell level with high spatial and temporal resolution3. In our laboratory, we investigate phenotypic variation in mycobacteria and strive to understand its bases and impact on the success of the tubercular pathogen. Given its importance in the physiology of M. tuberculosis4, we also aim to exploit it as a potential source of new weapons against tuberculosis. To this end, we integrate microsystems engineering with our core expertise in molecular microbiology and cell biology to develop devices that allow us to study the behavior of single cells under tightly controlled environmental conditions. This type of research is particularly demanding, especially working with a slow-growing pathogen that needs to be manipulated under biosafety level 3 conditions. However, we believe that single-cell approaches are essential to broaden the horizon on bacterial cell physiology and persistent subpopulations that are obscured by whole-population approaches. As we have shown with our work, quantitative single-cell approaches provide original insights that support the conception of original working hypotheses and the development of more effective antimicrobial strategies.
Indeed, conventional drug discovery models overlook microbial phenotypic variation, a critical factor in treatment failure that also leads to the emergence of drug resistance5. Recognizing this gap, we developed the µDeSCRiPTor strategy, which stands for microfluidic dynamic single-cell screening for pheno-tuning compounds (PTC). We leveraged our multidisciplinary expertise to develop a drug discovery model that incorporates phenotypic variation into the equation and aims to increase the susceptibility of the bacterial population to existing treatments. Central to our approach was the concept of “pheno-tuning” – a strategy that aims to reduce phenotypic variation from cell to cell in order to achieve a more homogeneous state of vulnerability across the bacterial population. This was made possible by our earlier single-cell study of the SOS DNA-damage response and the identification of a precise phenotypic state associated with drug susceptibility6. In short, by identifying compounds capable of tuning phenotypic variation, we aimed to improve the efficacy of existing anti-tubercular drugs, paving the way for more effective treatments (Figure 1). However, to achieve our goal, we first had to develop a customized multi-condition microfluidic platform (Figure 2), allowing us to capture subtle but crucial variations at the single-cell level while exposing growing microcolonies to candidate PTC. Thanks to this device, we carried out an unconventional drug screening and identified four PTC that triggered a homogeneous stress state in the bacterial population. We were able to determine this phenotypic change by using a fluorescent reporter strain of the SOS response and quantitative time-lapse microscopy. The next steps involved characterizing the cause of this phenotypic change, and determining whether this more homogeneous state of vulnerability synergizes with the activity of other drugs.
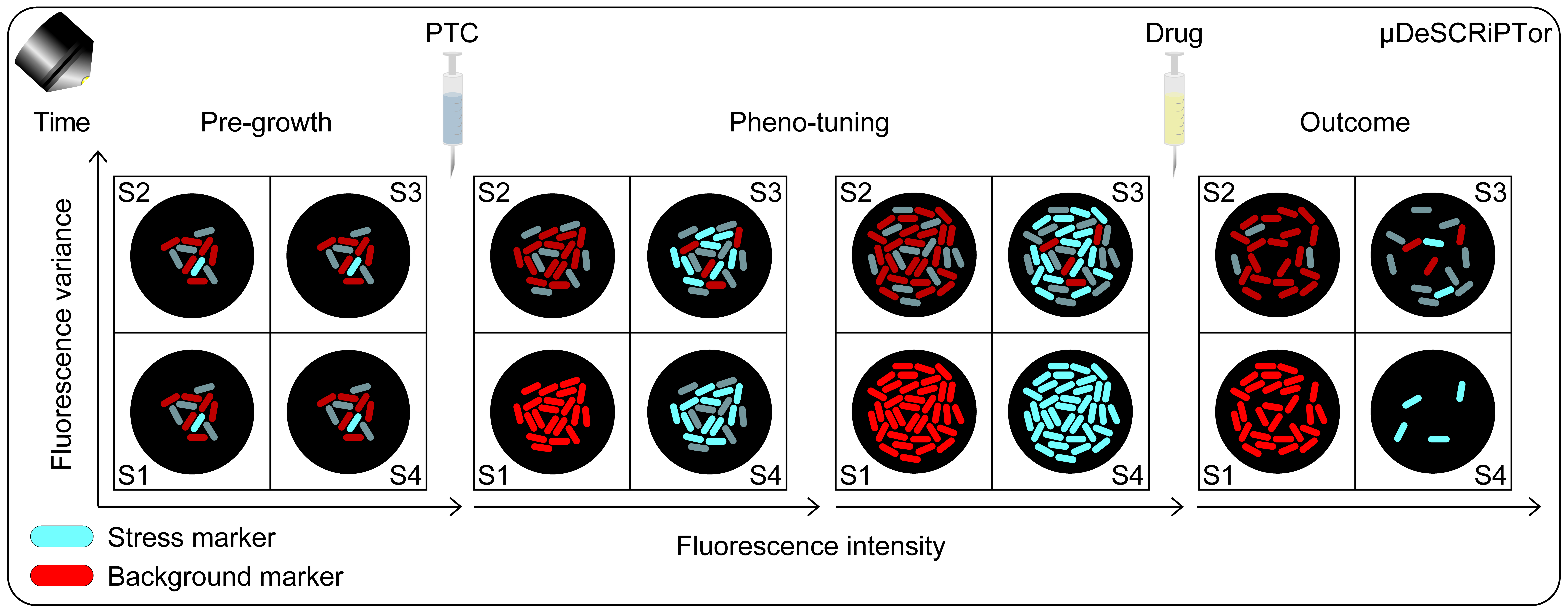
Figure 1. Schematic of the µDeSCRiPTor rationale. A fluorescent reporter strain of the SOS response6, as a stress marker (cyan), is combined with a constitutive background marker (red) and shows high levels of cell-to-cell variation under optimal growth conditions (pre-growth). Single-cell dynamics are captured by time-lapse microfluidic fluorescence microscopy through different experimental stages over time: pre-growth in the absence of stress; exposure to PTC; exposure to an anti-tubercular drug. Four possible scenarios are illustrated, relative to changes in the SOS fluorescence intensity and cell-to-cell variation: S1 represents low fluorescence and low variance; S2 represents low fluorescence and high variance; S3 represents high fluorescence and high variance; and S4 represents high fluorescence and low variance. An ideal PTC induces high levels of the stress marker and reduces the cell-to-cell variation according to S4. This scenario is expected to contribute to the best treatment outcome in combination with other anti-tubercular drugs.
Our journey was fraught with challenges related to microfabrication, optimization of the experimental setup, analysis of large amounts of microscopy datasets and multi-scale follow-up investigation. However, we were not deterred by the obstacles that stood in our way, and collaboration was the actual key to our success. From medicinal chemists who shared their compounds and expertise, to biostatisticians and bioimage analysts who disentangled various data sets, to biochemists who provided insights into the complex biology of M. tuberculosis, every member of our interdisciplinary team played a critical role in our shared goal of improving the treatment of tuberculosis. We were constantly driven by the fact that our efforts could one day improve tuberculosis patients’ lives. The turning point was the identification of a promising PTC hit, called M06. This heterocyclic organic molecule, derived from a phenanthroline scaffold, showed potent activity against M. tuberculosis and no citotoxicity. To explore the mode of action of M06, we integrated various omics, molecular, microbiological and biochemical approaches. We uncovered a multifactorial mechanism that impairs key cellular processes, including DNA replication and cell envelope stability, and significantly increases oxidative stress. Overall, our results confirm that exposure to the PTC M06 induces a homogeneous phenotypic shift, which undermines mycobacterial cells. Importantly, we have also shown that M06 potentiates the activity of existing anti-tubercular drugs, offering new hope in the fight against this resilient pathogen. The potential impact of our discovery is profound and promises to change the landscape of tuberculosis therapeutics by shortening the duration of treatment and preventing relapse of the disease.
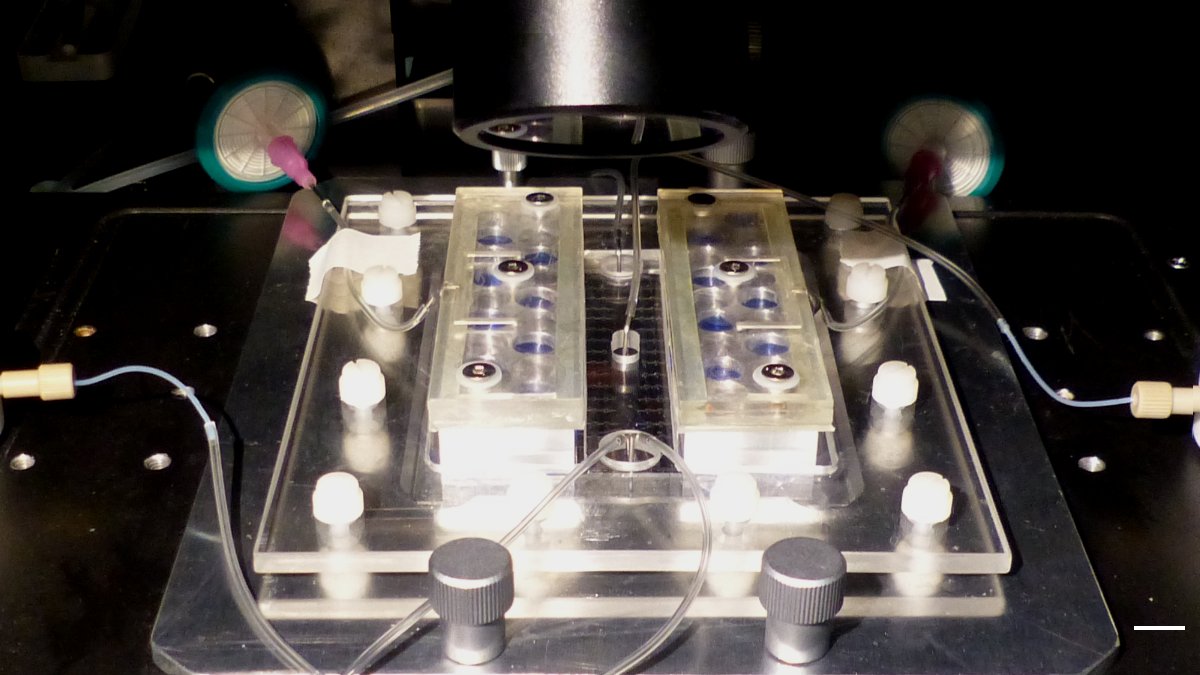
Figure 2. Picture of the multi-condition platform assembly on the wide-field fluorescence microscope stage. Scale bar = 1 cm.
Our journey continues as we will further explore the mechanisms underlying M06-mediated pheno-tuning through additional structural and biochemical analyses. Furthermore, the ongoing preclinical profiling of the safety and efficacy of M06 attests to our commitment to pursue its translation into clinical research. Our work extends beyond tuberculosis, as it provides tools and insights for combating other bacterial pathogens by targeting phenotypic variation. By scaling up our µDeSCRiPTor approach and improving single-cell analysis pipelines, we aim to support the broader microbiology community in the fight against infectious diseases.
In summary, our journey through µDeSCRiPTor exemplifies the power of interdisciplinary collaboration and innovative thinking in tackling the growing challenges posed by microbial pathogens, in this particular case M. tuberculosis. Our research also demonstrates the importance of considering phenotypic variation in the development of antimicrobial strategies and lays the foundation for future breakthroughs in microbial therapeutics.
REFERENCES
- Dartois, V. A. & Rubin, E. J. Anti-tuberculosis treatment strategies and drug development: challenges and priorities. Rev. Microbi ol. 20, 685–701 (2022).
- Dewachter, L., Fauvart, M. & Michiels, J. Bacterial heterogeneity and antibiotic survival: Understanding and combatting persistence an d heteroresistance. Cell 76, 255–267 (2019).
- Verstraete, L., Van den Bergh, B., Verstraeten, N. & Michiels, J. Ecology and evolution of antibiotic persistence. Trends Microbiol. 30, 466–479 (2022).
- Chung, E. S., Johnson, W. C. & Aldridge, B. B. Types and functions of heterogeneity in mycobacteria. Rev. Microbiol. 20, 529–5 41 (2022).
- Huemer, M., Mairpady Shambat, S., Brugger, S. D. & Zinkernagel, A. S. Antibiotic resistance and persistence—Implications for human health and treatment perspectives. EMBO Rep. 21, e51034 (2020).
- Manina, G., Griego, A., Singh, L. K., McKinney, J. D. & Dhar, N. Preexisting variation in DNA damage response predicts the fate of single mycobacteria under stress. EMBO J. 38, e101876 (2019).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in