Digging deeper into the individuality of bacteria
Published in Microbiology

We live in the age of single-cell microbiology. Fundamental processes that drive bacterial function, once accessible only through bulk methods that involve mixing millions of cells, can now be readily followed in a single cell. And when doing so, one is immediately struck by how different individual cells are from each other, in every aspect of their behavior. The common narrative for bacterial individuality is that it reflects the inherent stochasticity (randomness) of intracellular processes, which becomes amplified to the whole cell-level, making the behavior of individual cells unpredictable.
Representing single-cell phenotype as random has proven to be a useful abstraction. For example, in the study of gene expression, the measured cell-to-cell variability, described by the histogram of mRNA copy number, can often be reproduced using a simple stochastic model of transcription. However, the probabilistic picture also carries the danger of legitimizing our ignorance, by categorizing what is unknown to us as “random”, and therefore unknowable. But, in biology, what is unknown today may become knowable tomorrow, as technical innovations continuously enhance our ability to probe the inner working of cells.
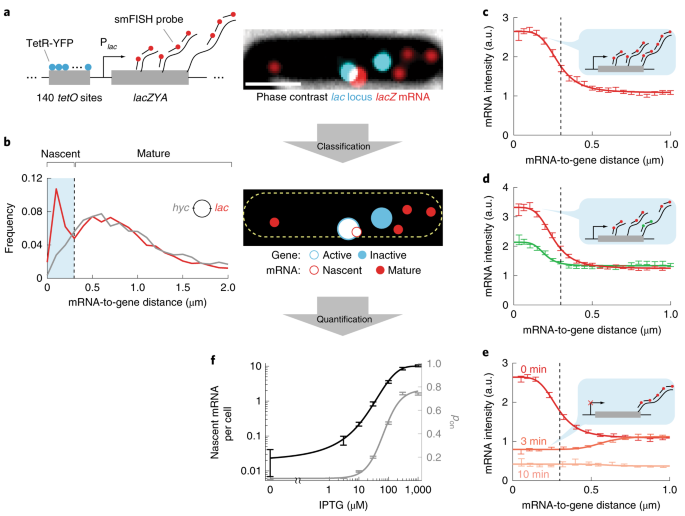
In our new paper, we try to demonstrate this point. By examining transcription as it takes place at an individual copy of a single gene in the cell, rather than making deductions from the overall number of mRNA molecules present in the cell, we found that transcription of a weakly expressed gene exhibits a transient pulse of activity around the time of gene replication. This temporal behavior could create strong cell-to-cell differences in the level of gene expression, because individual cells in the growing population are unsynchronized in their cell-cycle phase. We also found that two copies of the same gene, present in the cell as part of the replication cycle of the bacterial chromosome, may be highly correlated in their transcriptional activity, switching “on” and “off” together. This, too, is expected to increase cellular heterogeneity in a way that was previously unappreciated. The apparent coupling of gene transcription to other cellular events—gene replication and the activity of a sister gene copy—provides an example for additional drivers of bacterial individuality in gene expression, drivers that were hidden in earlier analyses.
Our paper: Mengyu Wang, Jing Zhang, Heng Xu, and Ido Golding, “Measuring transcription at a single gene copy reveals hidden drivers of bacterial individuality”, Nature Microbiology 2019.
Visit our lab at https://bacteriophysics.web.illinois.edu/
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026
Latest Content
Why is Singapore Identified in Global Research as Number One? How Physical Activity and Education Excellence Created a Global Leader




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in