Gut-brain axis in honey bees
Published in Microbiology

New insights into host-microbe interactions have drawn unprecedented attention to the microbial world in animals, both invertebrates and vertebrates. Gut microbiota plays an essential role in brain physiology and behaviors. Due to the difficulty of maintaining gnotobiotic animals, it is challenging to elucidate the contribution of individual gut members.
Pioneering research has identified
To find out how gut microbiota modulates honey bee behaviors, Zhang et al. used antibiotics to perturb the gut microbiota of bees under field conditions. Hive phenotypes, including the numbers of developing larva and capped brood cells, were disturbed. The relative abundances of the four species of Lactobacillus Firm5 were most strongly affected by antibiotic exposure. These results suggest that gut microbiota may influence honey bee hive behaviors. The ability to discriminate and remember odors is critical for honeybee social behaviors. Using gnotobiotic bees reared in the lab,
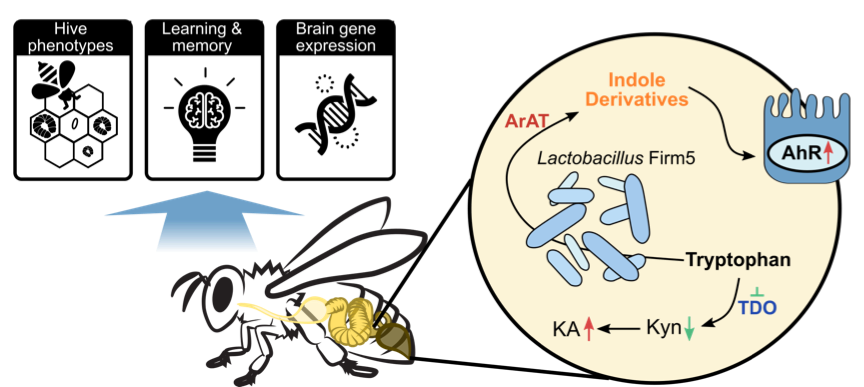
The study by Zhang et al. highlights the contributions of specific gut members to honeybee neurological processes, thus providing a promising model to investigate mechanisms of microbiota-gut-brain communication.
Original article: 10.1038/s41467-022-29760-0
Hao Zheng's Lab: zhenhaolab.org
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in