MORIARTY: A CRISPR tool to detect SARS-CoV-2
Published in Bioengineering & Biotechnology

It was June 2020 when the COVID-19 cases in the United States had crossed 2 Million with states like Florida reeling under immense pressure with escalating active cases1. Like other research laboratories across the globe, the lab headed by Prof. Hong Li based in the Institute of Molecular Biophysics, Florida State University (FSU) in Tallahassee, Florida went under lockdown to mitigate the spread of the virus. Many research projects including that on a CRISPR-Cas type III-A enzyme2 came to an abrupt halt. Under the frustration over COVID-19 and with our fresh knowledge of the type III-A enzyme (Csm), the idea to create a type III-A-based point-of-care testing for SARS-CoV-2 was born.
At the time, we were well aware of the previously described CRISPR-based virus detection technologies such as SHERLOCK3, HUDSON4, DETECTR5, HOLMES6,7 and many others that employ the single-subunit CRISPR-Cas systems like Cas9, Cas12 and Cas13. These pioneering studies laid the technical grounds to convert the viral nucleic acid-stimulated ribo- (RNase) or deoxyribo-nuclease (DNase) activities to detectable fluorescence or paper chromatography signals8. The type III-A enzyme we were studying has conceptually the same biochemical properties required for virus detection, but in addition, possesses unique dual detectable signals and internal signal amplification2. Though the idea of a tandem enzyme system has been explored with Cas13-Csm69,10, one with natural CRISPR-Cas systems was not yet available, owing to the complexity in enzyme production and unknown signalling conditions.
We were fortunate to have the availability of an all-in-one expression system for a Csm complex from our long-time collaborator, Prof. Michael Terns at the University of Georgia,2,11 that made it convenient to re-program and produce Csm in large quantity. Among the five subunits of Csm, Csm1 harbors both a DNase and an adenylate cyclase functionality upon viral RNA stimulation. The synthesized cyclic hexaadenylates (cA6) from ATP2 act as second messengers to allosterically activate another collateral RNase Csm612-14. Thus, a tandem Csm-Csm6 system programmed against SARS-Cov-2 RNA is capable of cleaving fluorescent DNA and RNA probes upon encountering the SARS-CoV-2 RNA present in patient swab RNA extracts (Fig. 1). Such a Csm-Csm6-based virus detection platform greatly benefits from the augmentability of the inherent dual collateral activities stemming from the Csm1 subunit and signal amplifiability by Csm6 via supplied oligonucleotides. We named this multifaceted, versatile but simple system: MORIARTY for Multipronged, One-pot, target RNA-Induced, Augmentable, Rapid, Test sYstem.
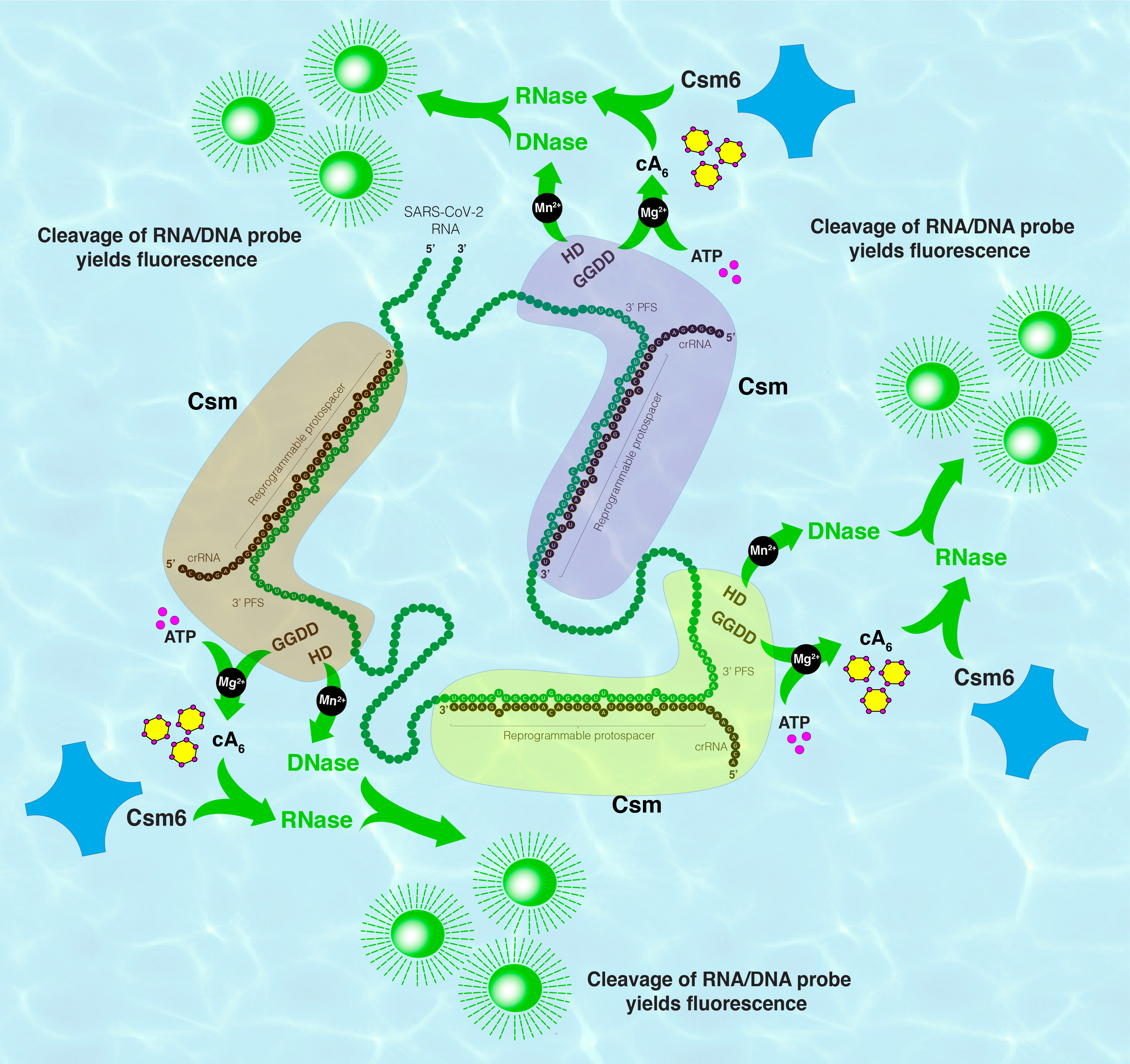
Figure 1: Schematic representation of COVID-19 diagnosis by amplification-free MORIARTY reaction assay. The presence of SARS-CoV-2 RNA leads to Csm-Csm6 activation causing cleavage of fluorescent DNA/RNA probes leading to a positive COVID-19 diagnosis.
Although MORIARTY’s design had excellent features and straightforward mechanics, harnessing the DNase & cyclase functionalities of Csm complex simultaneously was anything but easy! While in the presence of magnesium metal ions, Csm1 preferred to turn on its cA6 synthesis functionality, it contravened with the DNase functionality which preferred manganese metal ions. The inherent preference for either metal-ion condition was further complicated by the varied effects of reaction pH, salts, ATP, RNA/DNA fluorescent probes and the self-regulatory elements present in Csm or Csm6 on the two functionalities. The heroic efforts by our team systematically deciphered the behavior of MORIARTY under a multitude of conditions to arrive at the first model detection reaction at femtomolar sensitivity, still!
Viral titers vary greatly in infected patients15 that demand high sensitivity in detection. To further improve the detection limit of MORIARITY, we took two approaches. The first is to utilize multiplexing16 that enables a direct, or amplification-free, detection. We reprogrammed Csm to target three sections of the SARS-CoV-2 RNA encoding for the spike (S) protein and prepared the three Csm complexes. Under an optimized condition, the triplexing Csm-Csm6 system achieved a detection limit of 2000 copies/μL pre-quantified viral RNA.
The second approach utilized the natural ability of Csm to be activated by viral RNA transcription17 to pre-amplify the viral RNA. This strategy adopts isothermal amplification of viral RNA prior to detection through reverse transcription (RT) by T7 RNA polymerase and recombinase-polymerase amplification (RPA) that produce T7 promoter-fused DNA oligos3,18. Without the need for purification, the pre-amplified DNA oligos were readily detected by MORIARTY supplemented with T7 transcription components. The T7-MORIARTY reached an impressive sensitivity of 60 copies/μL pre-quantified viral RNA under 30 minutes (50 minutes in total including RT-RPA step).
Given the emergence of multiple variants of COVID-19 around the world19-21, we also explored the potential for MORIARTY to identify single nucleotide mutations in viral RNA. We found that, by pre-installing a mismatch between the guide RNA of Csm and the viral RNA, MORIARTY responded correctly to viral RNA with an additional mismatch. This paves the way for MORIARTY to be employed for distinguishing virus strains such as from the wild type to the Delta variants.
Through our collaboration with Prof. Jonathan H. Dennis at the Department of Biological Science at FSU, we accessed patient swab samples from FSU/Tallahassee Memorial Hospital COVID-19 testing center of varied and unknown viral titers and applied MORIARTY. For an unbiased, unhinged evaluation of MORIARTY’s applicability on patient samples, we established a double-blind study arrangement wherein Prof. Dennis would perform the standard q-RT-PCR detection procedure for human RNase P, SARS-CoV-2 N1-, N2- and E-genes, while we performed MORIARTY assays to detect S-gene, independently, on the same set of randomized samples. Without pre-amplification of viral RNA, MORIARTY correctly diagnosed COVID-19 status of patients with high titers or low q-RT-PCR Ct values. With T7-MORIARTY, we correctly diagnosed 7 out of the 8 q-RT-PCR-positive patients. These results are extremely encouraging, especially, those with the amplification-free mode. Without requiring sophisticated equipment or procedure, MORIARTY is poised to lower the testing costs and complexity in point-of-care testing.
MORIARTY is an off-the-shelf virus detection tool that is carefully optimized to exploit the Csm-Csm6 system with the future possibility of improved performance and additional functionality. During the conception and production of this work, other groups have envisioned their own type III CRISPR-based virus detection platforms22-24 that reflect the growing significance of class 1 CRISPR-effectors towards virus diagnosis among other biotechnological applications.
References:
1 AJMC Staff. A Timeline of COVID-19 Developments in 2020. doi:https://www.ajmc.com/view/a-timeline-of-covid19-developments-in-2020 (2021).
2 Sridhara, S. et al. Structure and Function of an in vivo Assembled Type III-A CRISPR-Cas Complex Reveal Critical Roles of Dynamics in Activity Control. bioRxiv, doi:https://doi.org/10.1101/2021.01.27.428455 (2021).
3 Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438-442, doi:10.1126/science.aam9321 (2017).
4 Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444-448, doi:10.1126/science.aas8836 (2018).
5 Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436-439, doi:10.1126/science.aar6245 (2018).
6 Li, S. Y. et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov 4, 20, doi:10.1038/s41421-018-0028-z (2018).
7 Li, S. Y. et al. Erratum: Author Correction: CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov 5, 17, doi:10.1038/s41421-019-0083-0 (2019).
8 Kaminski, M. M., Abudayyeh, O. O., Gootenberg, J. S., Zhang, F. & Collins, J. J. CRISPR-based diagnostics. Nat Biomed Eng 5, 643-656, doi:10.1038/s41551-021-00760-7 (2021).
9 Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439-444, doi:10.1126/science.aaq0179 (2018).
10 Liu, T. Y. et al. Accelerated RNA detection using tandem CRISPR nucleases. Nat Chem Biol 17, 982-988, doi:10.1038/s41589-021-00842-2 (2021).
11 Ichikawa, H. T. et al. Programmable type III-A CRISPR-Cas DNA targeting modules. PloS one 12, e0176221, doi:10.1371/journal.pone.0176221 (2017).
12 Jia, N. et al. Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity. Molecular cell 73, 264-277 e265, doi:10.1016/j.molcel.2018.11.007 (2019).
13 You, L. et al. Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference. Cell 176, 239-253 e216, doi:10.1016/j.cell.2018.10.052 (2019).
14 Niewoehner, O. et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543-548, doi:10.1038/nature23467 (2017).
15 Cevik, M. et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2, e13-e22, doi:10.1016/S2666-5247(20)30172-5 (2021).
16 Fozouni, P. et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184, 323-333 e329, doi:10.1016/j.cell.2020.12.001 (2021).
17 Liu, T. Y., Liu, J. J., Aditham, A. J., Nogales, E. & Doudna, J. A. Target preference of Type III-A CRISPR-Cas complexes at the transcription bubble. Nat Commun 10, 3001, doi:10.1038/s41467-019-10780-2 (2019).
18 Joung, J. et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N Engl J Med 383, 1492-1494, doi:10.1056/NEJMc2026172 (2020).
19 Tang, J. W., Toovey, O. T. R., Harvey, K. N. & Hui, D. D. S. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J Infect, doi:10.1016/j.jinf.2021.01.007 (2021).
20 Tang, J. W., Tambyah, P. A. & Hui, D. S. Emergence of a new SARS-CoV-2 variant in the UK. J Infect, doi:10.1016/j.jinf.2020.12.024 (2020).
21 Kirby, T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir Med, doi:10.1016/S2213-2600(21)00005-9 (2021).
22 Santiago-Frangos, A. et al. Intrinsic signal amplification by type III CRISPR-Cas systems provides a sequence-specific SARS-CoV-2 diagnostic. Cell Rep Med 2, 100319, doi:10.1016/j.xcrm.2021.100319 (2021).
23 Steens, J. A. et al. SCOPE enables type III CRISPR-Cas diagnostics using flexible targeting and stringent CARF ribonuclease activation. Nat Commun 12, 5033, doi:10.1038/s41467-021-25337-5 (2021).
24 Grüschow, S., Adamson, C. S. & White, M. F. Specificity and sensitivity of an RNA targeting type III CRISPR complex coupled with a NucC endonuclease effector. doi:https://doi.org/10.1101/2021.09.13.460032 (2021).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in