Non-invasive diagnosis and surveillance of bladder cancer with driver and passenger DNAmethylation in a prospective cohort study
Published in Cancer and Genetics & Genomics
The management of urothelial carcinoma, a type of bladder cancer, faces significant challenges, particularly in accurately diagnosing and classifying tumors. Bladder cancer can be classified into two main types: muscle-invasive bladder cancer (MIBC) and non-muscle-invasive bladder cancer (NMIBC). These classifications are crucial because they dictate very different treatment approaches.
Currently, bladder cancer is primarily diagnosed through histopathological assessment, which involves examining tissue samples under a microscope. This method is invasive, expensive, and can sometimes lead to complications such as infections. Additionally, the accuracy of this method can vary due to subjective interpretation by pathologists and the selection of tissue samples.
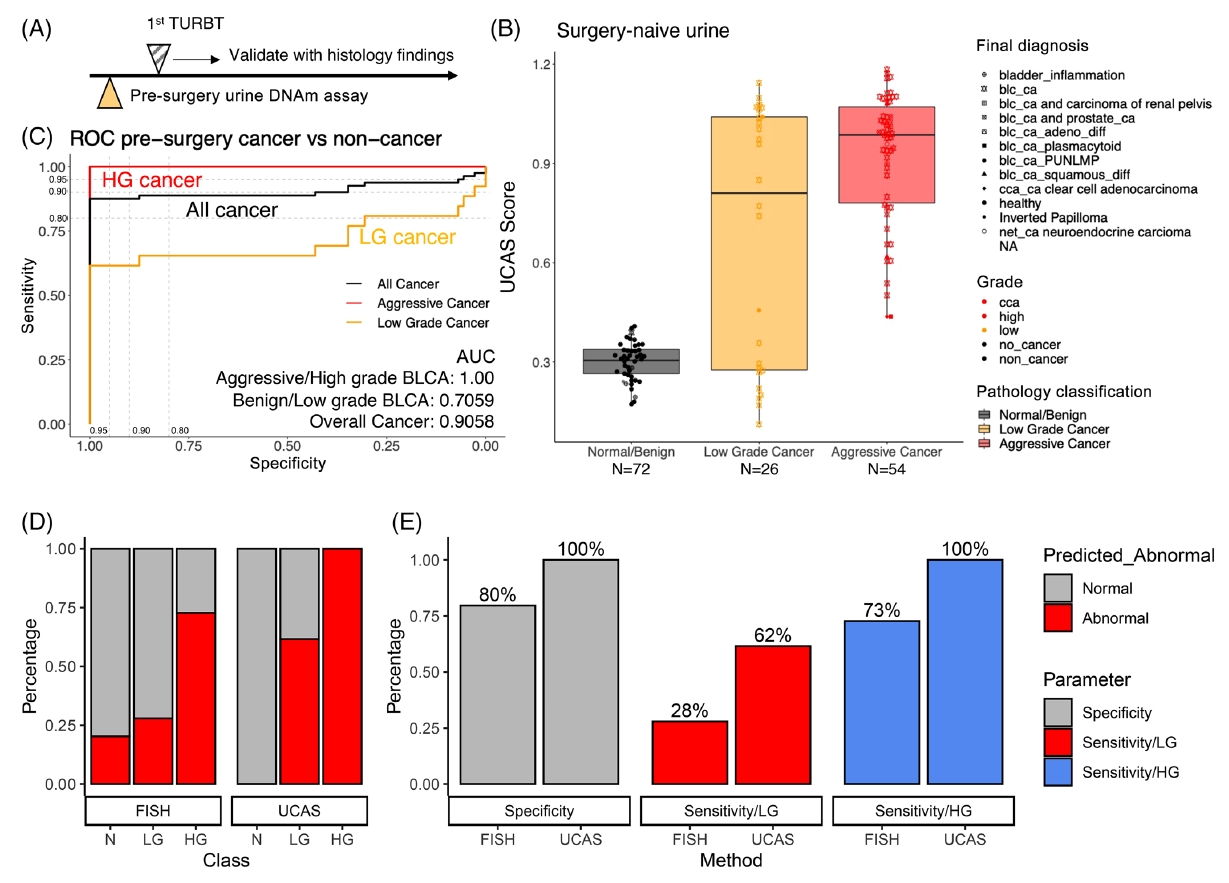
Bladder cancer staging, which determines how far the cancer has spread, is vital for treatment. Incorrect staging can lead to inadequate treatment or unnecessary surgery, both of which can significantly affect a patient’s quality of life. An alternative to tissue sampling is liquid biopsy, which uses urine samples to detect cancer. This method is less invasive and can identify markers like DNA, RNA, and proteins that are indicative of cancer. However, current urine-based tests have limitations in sensitivity and specificity, particularly in early detection and in distinguishing between different cancer subtypes.
Our previous study shows that DNA methylation, a genetic modification that does not change the DNA sequence but can affect gene activity, could be a promising marker for bladder cancer detection [1], analyzed by the tool named EpiTrace [2]. Different patterns of methylation can indicate the origin of the cancer cells or changes that occur during cancer development.
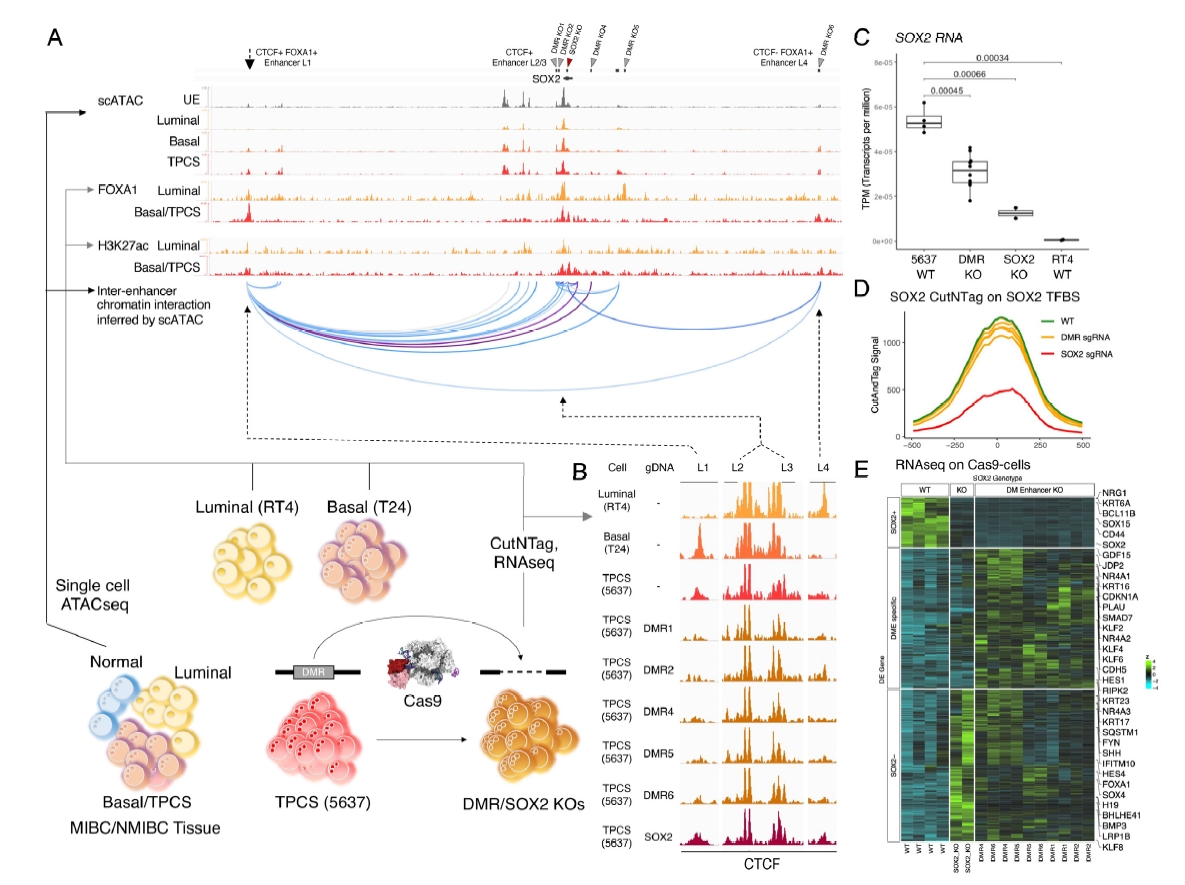
The current study focuses on using DNA methylation to improve the non-invasive detection and classification of bladder cancer through a new approach that combines advanced genetic analysis with statistical methods.
This approach aims to differentiate between methylation patterns that are inherent to cancer cells and those from non-cancer cells, which could lead to more accurate diagnosis and staging of bladder cancer. We tested this method in a large multi-center study involving 224 patients across China, showing promising results for its use in clinical practice.
References
[1] Xiao, Y. et al. Integrative Single Cell Atlas Revealed Intratumoral Heterogeneity Generation from an Adaptive Epigenetic Cell State in Human Bladder Urothelial Carcinoma. Adv Sci (Weinh), e2308438 (2024).
[2] Xiao Y., et al. Tracking single-cell evolution using clock-like chromatin accessibility loci. Nat Biotechnol, doi: 10.1038/s41587-024-02241-z. Epub ahead of print (2024).
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in