Precise post-transcriptional gene silencing with CRISPRδ
Published in Bioengineering & Biotechnology and Cell & Molecular Biology
The development of precise and effective methods to study biological functions and treat diseases is a perpetual pursuit in biological sciences. The field of gene expression is no exception where the introduction of CRISPR-Cas technology has played a major role, providing numerous tools for its manipulation.
In recent years, CRISPR-Cas13 systems have been introduced as tools for post-transcriptional gene silencing, due to their ability to selectively target and cleave RNA. These systems consist of a single type VI protein effector and a guide RNA that contains a hairpin region necessary for binding to the effector protein and a 20-30 nt spacer region that hybridizes with the target RNA. Given their high precision, tight-binding, and effective cleavage of the target RNA, Cas13 systems rapidly attracted attention as tools for targeted gene knockdown. However, the nonspecific cleavage of bystander RNAs upon Cas13’s activation, also known as collateral activity, has posed a barrier to the usage of Cas13 as a transcript-specific tool.
Interestingly, the introduction of mutations at the protein’s catalytic residues responsible for its RNase activity (target-specific and collateral) can render the protein catalytically inactive. Such “dead” or dCas13 proteins lose their RNA-cleavage ability but retain their RNA-specific and tight-binding ability. This intriguing feature of dCas13 proteins has been utilized for various applications (RNA editing, control of alternative splicing, and RNA tracking among others) and became our major hypothesis's basis. We reasoned that the placement of dCas13 on target transcripts could be used as a tool to block the translational machinery, resulting in target-specific translational repression. Such an approach could eventually bypass the collateral effect of Cas13 achieving high precision gene silencing.
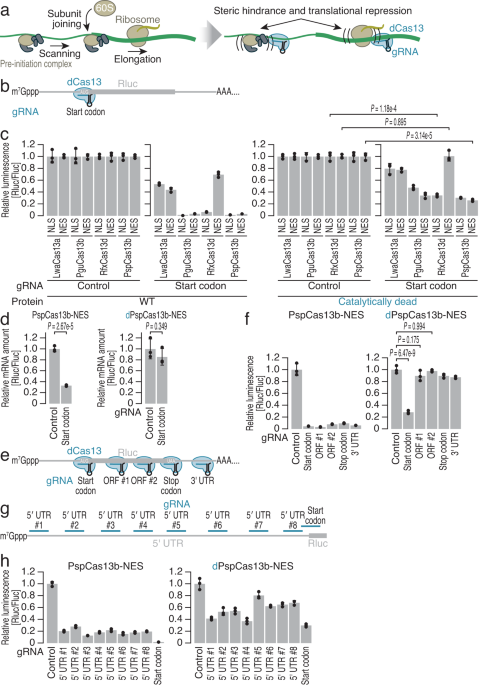
Setting out to test our hypothesis, we tried several different dCas13 proteins and also tested whether their subcellular localization could impact their performance. The view of the first luminescence signal reduction noting that dCas13 proteins could repress the translation of the target reporter transcript was indeed exciting. That initial excitement was followed by downstream tests that gave us more insight into how this new tool works. We realized that different dCas13 proteins and their subcellular localization showed different efficacy in translation repression. Importantly, we found that dCas13-induced translation repression was more efficient when blocking the pre-initiation complex rather than the fully assembled ribosome. Specifically, targeting the start codon of the target transcript provided more consistency in our results. Among all the tested proteins, we found the inactive version of Prevotella sp. P5-125 Cas13b (dPspCas13b) localized in the cytoplasm to have the most robust effect and chose it for downstream experiments. A major finding from these experiments was that dCas13-induced repression of translation did not degrade the target RNA. To understand how the gRNA properties and hybridization position affect the system’s efficacy, we tested variables such as gRNA length and position against the start codon. We observed that 19 to 25 nt long spacers as well as central positioning of the anti-start codon within the gRNA enhanced the efficacy of dCas13.
In our discussions on how to name our system, we decided to maintain CRISPR and distill dCas13’s function in the single letter δ (delta: DEpLetion of Translation by blockAde).
Accuracy has always been a major challenge in the development of gene silencing methods. In our work, to survey the specificity of CRISPRδ, we used a combination of next-generation sequencing methods including RNA sequencing and ribosome profiling. The latter is a method to determine protein synthesis by deep sequencing of ribosome-protected RNA fragments generated by ribonuclease digestion. This combination of methods confirmed the high specificity of CRISPRδ. Except for the target gene, we could not observe non-specific translation repression and also we did not observe RNA degradation of either the target or non-targeted transcripts.
Understanding that CRISPRδ’s repression efficiency was modest compared to other methods such as active Cas13 or RNAi, we searched for ways to optimize the system. Our efforts mainly focused on fusing the dCas13 protein with translational suppressor proteins. Those sets of experiments revealed that fusion with 4EHP protein offered the highest repression when compared to dCas13 protein alone.
In this study, we developed a new method to interfere with gene expression by blocking the translational machinery. Our results showed that CRISPRδ is a precise and versatile tool that can be used in different cell lines and repress transient as well as endogenous gene expression. An important feature of this study is that CRISPRδ successfully repressed not only canonical cap-dependent but also internal ribosome entry site (IRES)-mediated as well as repeat-associated non-AUG (RAN) translation. CRISPRδ’s translation repression effect is modest compared to other existing methods; however, further studies that focus particularly on the improvement of the system (e.g., enhancement of the stochastic expression of CRISPRδ) could decrease the current efficiency gap. Finally, we believe that the features of our system, including the high specificity and the retainment of the target RNA’s integrity will also aid in areas where traditional methods face limitations (e.g., deciphering the coding-independent functions of mRNAs).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in