
The self-assembly of amyloid proteins is a fascinating phenomenon that results in linear, nearly one-dimensional nano-sized fibers (AKA nano-fibrils) with striking biological and physical characteristics. The process of fibrillar protein self-assembly is, on the one hand, linked to the development of more than 60 disorders, among them Alzheimer’s and Parkinson’s diseases and Type II diabetes. On the other hand, nature is filled with fibrillar materials with exceptional properties in terms of mechanics, piezoelectricity, and sensitivity to changes in the chemical environment and more.
My research group is interested in understanding the driving forces that bring protein molecules together to form nanoscale structures and how changes to these processes affect the material characteristics of the resulting constructs. Intrigued by the ability to tune the mechanical properties of silk fibers by changing, for example, the level of water content, applied shear and chemical environment, we decided to “zoom in” on the molecular changes that occur in local protein-protein interactions that affect silk self-assembly, in general, and its material characteristics, in particular.
To this end, we generated peptide models that mimic the repetitive sequence of the silk hydrophobic motif, GAGAGS. Of note, such rich-in-G-and-A amino acid sequences are found in proteins whose fibrillation is associated with the development of neurodegenerative disorders, including Huntington’s, Parkinson’s, mad cow diseases and many more. Thus, understanding the driving forces behind the fibrillation of proteins containing such sequences is valuable not only for the development of new artificial protein-based materials but also for obtaining new insight into the molecular mechanisms of disease development.
We introduced steric constrains into the peptide assembly by replacing amino acid G with the aromatic amino acid F, Y or W. These three aromatic amino acids differ in their hydrophobicity and ability to form hydrogen (H) bonds. For example, while F is purely hydrophobic, Y can serve as a strong H donor.
We observed that low F and Y content (up to 2%) does not severely affect the peptide’s self-assembly process. The IR analysis confirmed the presence of amyloidogenic ß-sheet conformations, and the electron diffraction analysis exhibited inter-sheet and inter-strain distance values characteristic of classical amyloids. Interestingly, the presence of W (2%) suppressed the peptide self-assembly process.
When we increased the fraction of hydrophobic amino acids (up to 30%) in the peptide sequence, we observed pronounced differences in the nanomechanics of the peptide assemblies. However, the structural analysis (IR, electron diffraction) did not show dramatic changes in peptide conformation and structure.
Frustrated, we decided to look into the process of peptide fibrillation and, in particular, into the H-bonded network. Strikingly, our analysis of the chemical kinetics of peptide self-assembly revealed notable differences in the nucleation time of the various peptide constructs. The longest time was observed for W-containing peptides, which we explained by a solvation effect that contributed to the metastability of the W-containing peptide complex.
Next, we performed an X-ray photoelectron spectroscopy (XPS) analysis. This is quite an unconventional approach for H-bonded network analysis, since the only element that is invisible for XPS is hydrogen. However, we tracked the changes in the binding energies of the N and O atoms involved in H-bonded network formation. This led us to uncover pronounced differences in the H-bonded network of the peptide assemblies containing a high fraction of either F, Y or W. These differences were in line with the variations in the nanomechanics. For example, the softer materials contained a lower fraction of H-bonds, while stiffer materials demonstrated increased H-bond content.
Thus, balancing parameters such as solvation, H-bonding and the kinetics of protein fibrillation can be used as a new approach for material design via controlling the fibrillar self-assembly process.
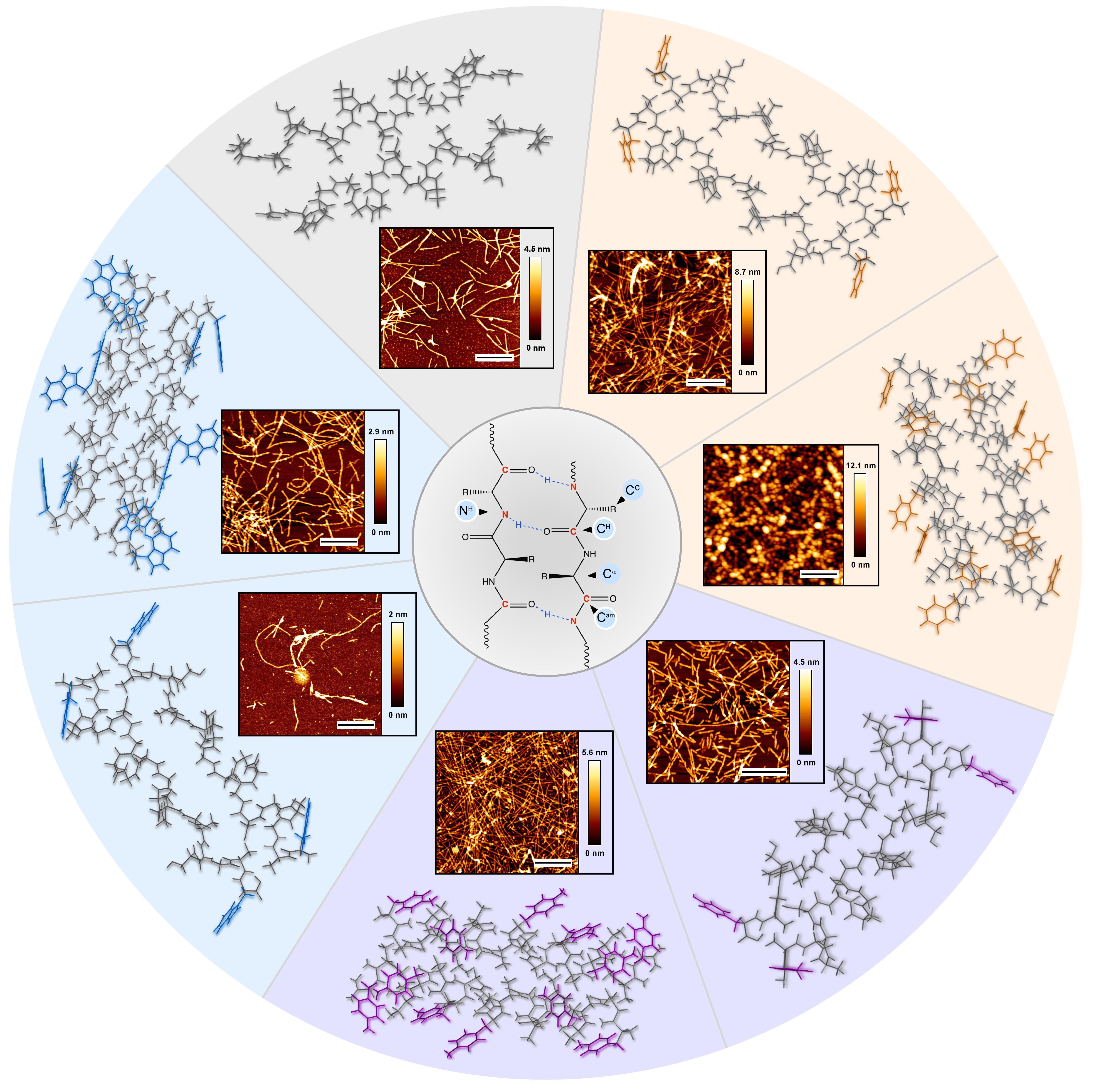
Schematic representation of the peptide design approach enabling modulation of H-bonded network and fine-tuning the mechanics of the peptide assemblies. Figure showing (from center towards the circle edges) the scheme of the peptide H-bonded network, atomic force microscopy images of the peptide assemblies and the relative orientation of the two peptide molecules constructing these assemblies.
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
f-block chemistry
Publishing Model: Open Access
Deadline: Feb 28, 2026
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in