Replication-competent HIV-guided CRISPR/Cas9 approach reveals antiviral factors and mechanisms
Published in Microbiology, General & Internal Medicine, and Immunology

Human cells express an astounding variety of proteins that may help to protect us against viral pathogens. Most of the best characterized antiviral proteins, called restriction factors, share common properties, such as they are induced by interferons, interact with viral components, and show evidence for positive selection due to the ever-ongoing evolutionary arms race between host and invading viral pathogens. We have long been interested in the discovery of such antiviral factors, especially against HIV-1, the causative agent of AIDS. In the recent decades a plethora of work from various laboratories has uncovered a very diverse set of restriction factors. However, it remained painfully evident that important antiviral factors that play roles in HIV-1 transmission and/or are counteracted by the viral accessory Vif, Vpr, Vpu or Nef proteins are not yet known. Finding these ‘missing restriction factors’ is a challenging (and sometimes frustrating) task since antiviral factors are structurally and functionally highly diverse and those that remain to be discovered may not share the common properties mentioned above.
The present project was thus driven by the vision to coerce HIV-1 to reveal its cellular antagonists, and thus unravel what constitutes the first line of defenses against viruses. To achieve this, two highly talented and persistent PhD students Caterina Prelli Bozzo and Alexandre Laliberte developed advanced methodologies to equip HIV-1 with guide RNAs (gRNAs) that target potential restriction factors. In a nutshell, the basic idea was that HIV-1 constructs expressing single gRNAs inactivating antiviral genes will have a selection advantage during cell culture passage. Usually, the evolutionary, emergence of “the fittest” approaches do not reveal the mechanism(s) underlying the selective advantage. Here, however, the guide RNAs acts as a barcode for the advantage, revealing the identity of the targeted genes by simple sequencing (Fig. 1). Notably, the gRNAs can only confer a selection advantage to HIV-1 in the presence of artificial expression of the bacterial Cas9 endonuclease, thus avoiding biosafety issues. Thus, the HIV-CRISPR/Cas9 system is not functional in human or mammalian cells, except when they have been specifically modified to express Cas9.
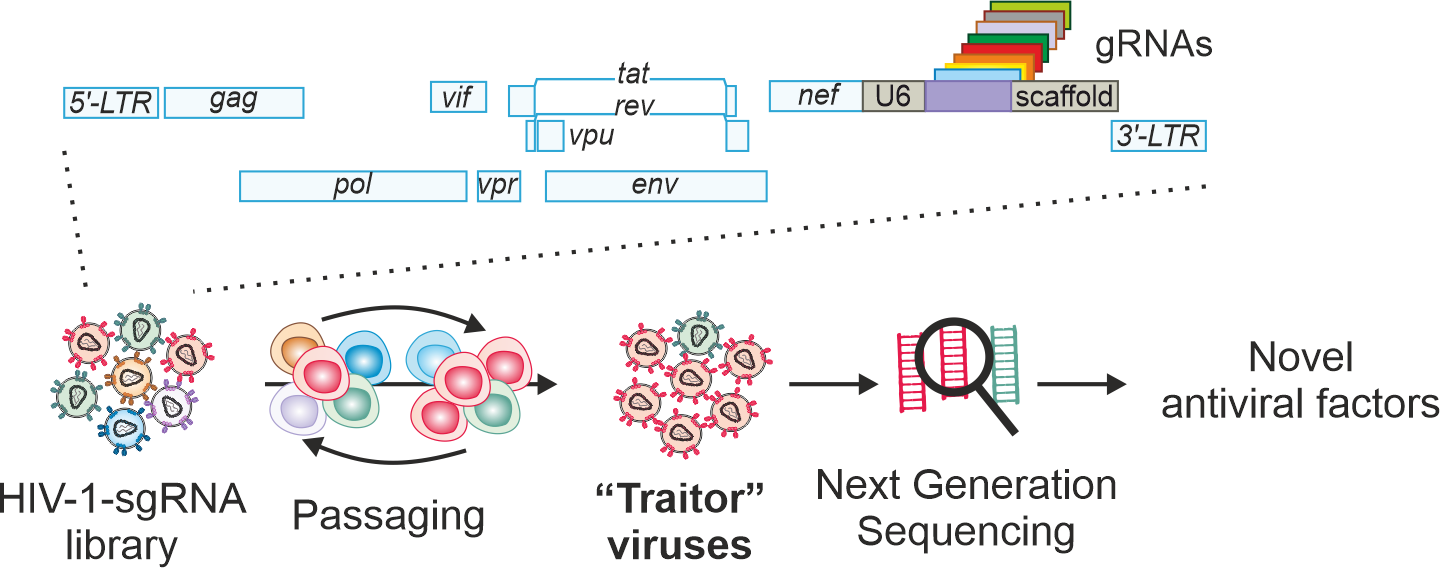
Fig. 1. Outline of the CRISPR/Cas9-based virus-guided discovery approach. Proviral HIV-1 constructs are engineered to contain the sgRNAs expression cassette between the nef gene and the 3`LTR. Libraries of HIV-1 sgRNA viruses are passaged every two days in Cas9-expressing cells. The frequencies of HIV-1 sgRNAs are determined by next-generation sequencing of the supernatant every two days. Target genes of sgRNAs that are selected and hence are associated with an advantage for viral replication are cloned and examined for their antiviral activity and mechanism. Genome size is not to scale.
While our method, termed “Traitor virus” approach, seems straight-forward, we had to overcome several obstacles during the earlier stages of the project. For example, proviral HIV-1 constructs are flanked by repetitive sequences, long terminal repeats (LTRs), that are notorious for their ability to recombine. Hence introducing the gRNA expression cassette into the proviral genome required careful application and optimization of cloning techniques. In addition, in the HIV-1 genome the information is densely packed and genes and functional elements overlap. To allow insertion of a cassette coding gRNAs while maintaining expression of all viral proteins, we separated the nef gene from the 3`LTR (Fig. 1). Eventually, after promising proof of concept studies using HIV-1 constructs expressing tetherin and GBP5, two well-known restriction factors, we engineered libraries of >1500 replication-competent HIV-1 constructs each expressing a single sgRNAs to target >500 cellular genes using elegant and adaptable single-step reactions. Joining forces with the group of Helmut Blum at the Gene Center in Munich, we set out to identify novel restriction factors by passaging the complex libraries of HIV-1-sgRNA viruses on Cas9 expressing T cells. Unfortunately, gRNA expression cassettes were rapidly lost during passage of these fully replication-competent HIV-1 constructs in cell culture. By elimination repetitive hot-spots of recombination within the nef gene without changing its coding sequences, we fixed this problem and managed to generate replication-competent and stable HIV-1 sgRNA libraries.
Now ready to start the exciting part of the project, Alex and Caterina passaged the HIV-1 gRNA libraries in CD4+ cells lines previously engineered to express Cas9. Indeed, passaging of the viral libraries robustly enriched HIV-1 encoding sgRNAs against certain cellular genes frequently by more than two orders of magnitude (Fig. 2a). To ensure the relevance of our findings, we generated the analogous library in the context of a transmitted-founder HIV-1 strain and performed the passaging in two different cell lines, CEM-M7 and SupT1 cells. In addition, functional analyses in primary human cells support that the HIV-driven CRISPR screen robustly identified restriction factors targeting virus transcription, release and infectivity (Fig. 2b).
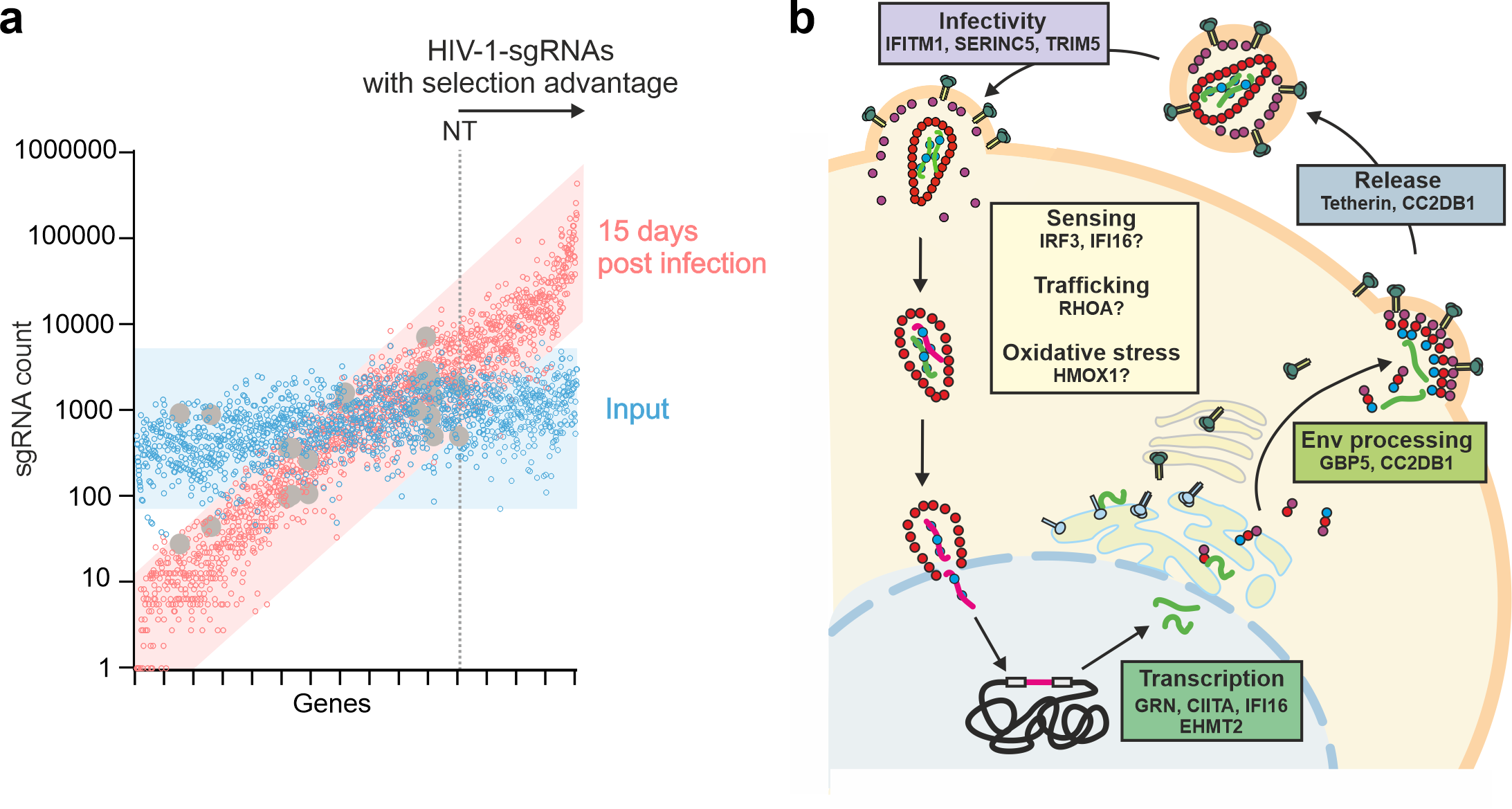
Fig. 2: a, The composition of the HIV-1 sgRNA library changes over the course of 15 days of replication in T cells and sgRNAs that convey a replication advantage get enriched. Scatter plot of individual sgRNA counts in CEM-M7 cells supernatants 15 days post-infection (red outlines) and in the input (blue outlines) versus gene names sorted by fold enrichment. NT and dotted line indicate the occurrence of the first non-targeting control sgRNA. sgRNAs that convey a replication advantage are right of the dotted line. b, Schematic overview of the HIV-1 replication cycle and factors detected in the TV-driven screening approach.
Pandemic HIV-1 strains are well adapted to humans and their accessory proteins counteract major human restriction factors. Thus, we hypothesized that lack of accessory viral genes will increase selection for sgRNAs targeting antiviral factors that are otherwise counteracted by the corresponding accessory factors. Indeed, lack of the nef gene not only increased selection of sgRNA viral constructs targeting the established Nef target SERINC5 (that otherwise impairs virion infectivity) but also IFI16 that sequesters the transcription factor Sp1. Functional analyses of a variety wild-type and nef-defective clade B and C HIV-1 strains confirmed that Nef reduces viral sensitivity to restriction by IFI16.
The sgRNAs function essentially like artificial accessory viral genes – with the important difference that they reveal their cellular targets. Thus, future directions are to expand the libraries to targeting genes genome-wide and to use HIV-1 with defined genetic defects to discover further targets of Vif, Vpr, Vpu and Nef or poorly adapted SIVcpz strains to define adaptations required for successful zoonotic transmission. The high versality of our virus-guided approach will reveal restrictive mechanisms in different cell types and in the presence of various kinds of selection pressures, such as antiviral interferons and cytokines. In addition, our approach also selects for sgRNA targeting cellular pathways or features rendering cells less permissive for HIV-1 replication.
Our approach and results are relevant beyond HIV/AIDS because innate defense mechanisms evolved to provide broad protection against a wide range of viral pathogens. Although our current HIV-1 gRNA libraries and Cas9-expressing cell lines present many exciting opportunities to explore virus-host interactions, challenges remain. Perhaps most importantly establishing approaches for selection in Cas9 expressing primary T cells. Nonetheless, our current analysis of HIV-1 gRNA libraries in Cas9-expressing cell lines already identified restriction factors targeting viral entry, sensing, transcription, release and infectivity and thus offers many exciting perspectives for further investigation.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Ask the Editor - Immunology, Pathogenesis, Inflammation and Innate Immunity
Got a question for the editor about the complement system in health and disease? Ask it here!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in