Tiny Messengers: How Worms Teach Us About Human Diseases
Published in Neuroscience, Cell & Molecular Biology, and Genetics & Genomics
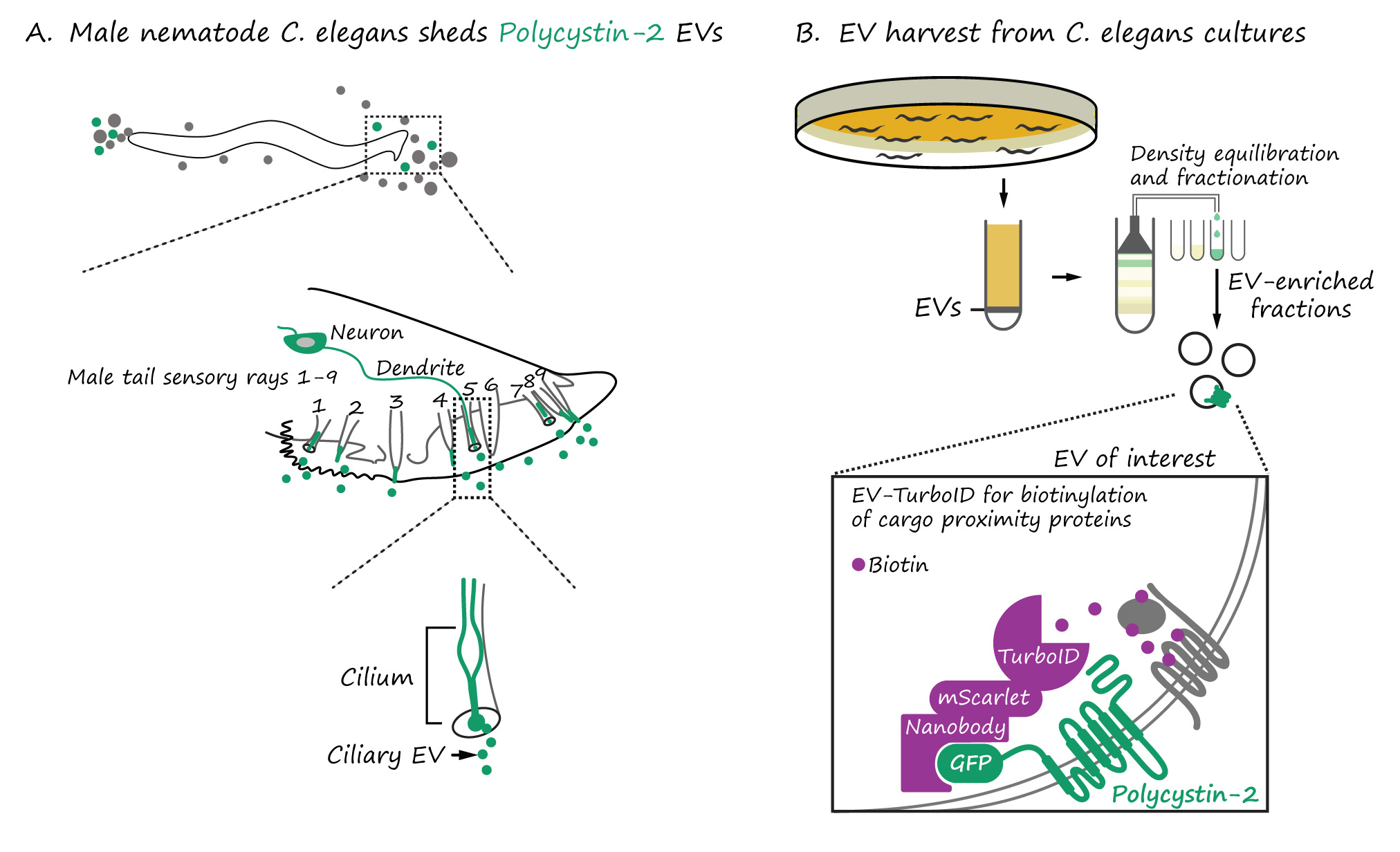
Our scientific project began with an art-worthy observation – an explosive release of extracellular vesicles (EVs) from C. elegans “nose” and tail sensory neurons. Each EV carries GFP-labeled polycystin-2, a protein that in humans is linked to devastating polycystic kidney disease and is a cargo of human urinary EVs.

In worms, these polycystin EVs bud into the environment from the tip of the neuronal sensory cilium, a tiny ancient hair-like antenna. The cilium is no ordinary cell projection. It’s a gated, energy-hungry compartment with specific ion channels, GTPases, and transport machinery. If the cell invests so heavily into regulating what enters and exits the cilium, then surely the EVs it sheds are not accidental. Could these membrane-bound particles be messengers with meaningful signaling roles?
This question became especially compelling when we considered that these EVs originate from neurons - cells associated with memory, learning, neuronal signaling, and spatial-temporal encoding. Could ciliary EVs contribute to some of these elusive neuronal functions? The hypothesis felt big—but so was the challenge: how does one even begin to trace the function of specific ciliary EVs?
The EV Identity Crisis
EV research is exploding. Studies have linked EVs to everything from tissue regeneration after injury to the spread of pathological factors in disease. However, most of this work analyzes mixed EV populations harvested from complex fluids - blood, urine, or culture media - where all vesicle types are lumped together. It’s like trying to deduce the purpose of a single letter by sorting through an entire city’s mailroom: everything is there, and nothing is specific.
What’s missing is resolution. The field has long lacked tools to precisely define the cargo of specific EV subtypes—especially those released from small and spatially constrained compartments like the cilium. Understanding these cargoes could shed light on the pathophysiology of ciliopathies, including polycystic kidney disease.
A Rosetta Stone of Biology
To solve a biological mystery, you need the right model. For us, that model was Caenorhabditis elegans, a transparent, millimeter-long worm. Why worms? Think of them as the Rosetta Stone of biology. Just as the original Rosetta Stone allowed linguists to decipher ancient hieroglyphs by presenting the same text in multiple languages, C. elegans offers a uniquely decipherable biological system. It is genetically tractable, easy to image, and remarkably conserved at the molecular level.
And crucially: C. elegans sheds its ciliary EVs directly into the environment. That meant we could culture millions of worms, isolate the EVs, and dissect their content—something nearly impossible in mice, which don’t produce enough urine to recover sufficient polycystin EVs for analysis.
What’s inscrutable in mice and humans - especially something as heterogeneous and elusive as EVs - becomes visible and manageable in worms.
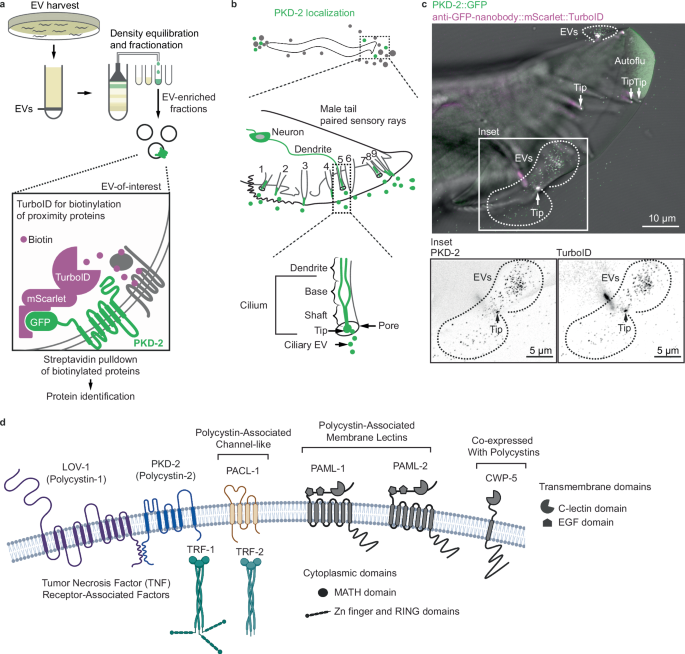
A targeted method to identify EV cargo: EV-TurboID
In 2022, we showed that C. elegans EVs contain RNA and diverse protein cargo. But we couldn’t tell which tissues contributed which cargo, and which protein were truly co-packaged in polycystin EVs.
To solve this, we developed EV-TurboID, a modular proximity labeling system in which an engineered biotin ligase (TurboID) is tethered to polycystin-2 via a nanobody. This enzyme tags proteins that linger near polycystin-2, generating a high-resolution “proximity map” of the polycystin EV cargo.
From Thousands to Eight
Our earlier attempts at in-bulk identifications yielded nearly 3,000 EV candidate proteins, whereas EV-TurboID narrowed the list to eight. Using CRISPR/Cas9 technology to generate endogenous fluorescent reporters for each candidate, we observed their behavior in real time in living animals.
Among the top hits were transmembrane C-type lectins, a channel-like protein, and a family of TRAF adaptors best known for relaying immune and growth factor signals in human cells. TRAFs had never before been seen in cilia, and we observed them not only inside the ciliary shaft, but also at the tip and inside EVs – accompanying the polycystin complex.
That wasn’t all. TRAF proteins, we found, didn’t just randomly drift into EVs. Their loading was conditional - dependent on polycystin-1 (LOV-1). Without LOV-1, they entered the cilium but were excluded from the tip and EVs. It was as if LOV-1 served as a molecular bouncer permitting entry into the vesicular club only when present.
Function, Not Flotsam
Our findings challenged a long-standing assumption that EVs are a passive means of waste disposal. Instead, we found that EV cargo is curated, and loading is regulated where LOV-1 acts as a molecular organizer, coordinating cargo recruitment and departure via EVs.
The implications are significant. In polycystic kidney disease, mutations in polycystin-1 are often more severe than those in polycystin-2. Our findings suggest that this may be because, without polycystin-1, EVs are still released—but their signaling payload is incomplete or incorrect, potentially contributing to disease progression.
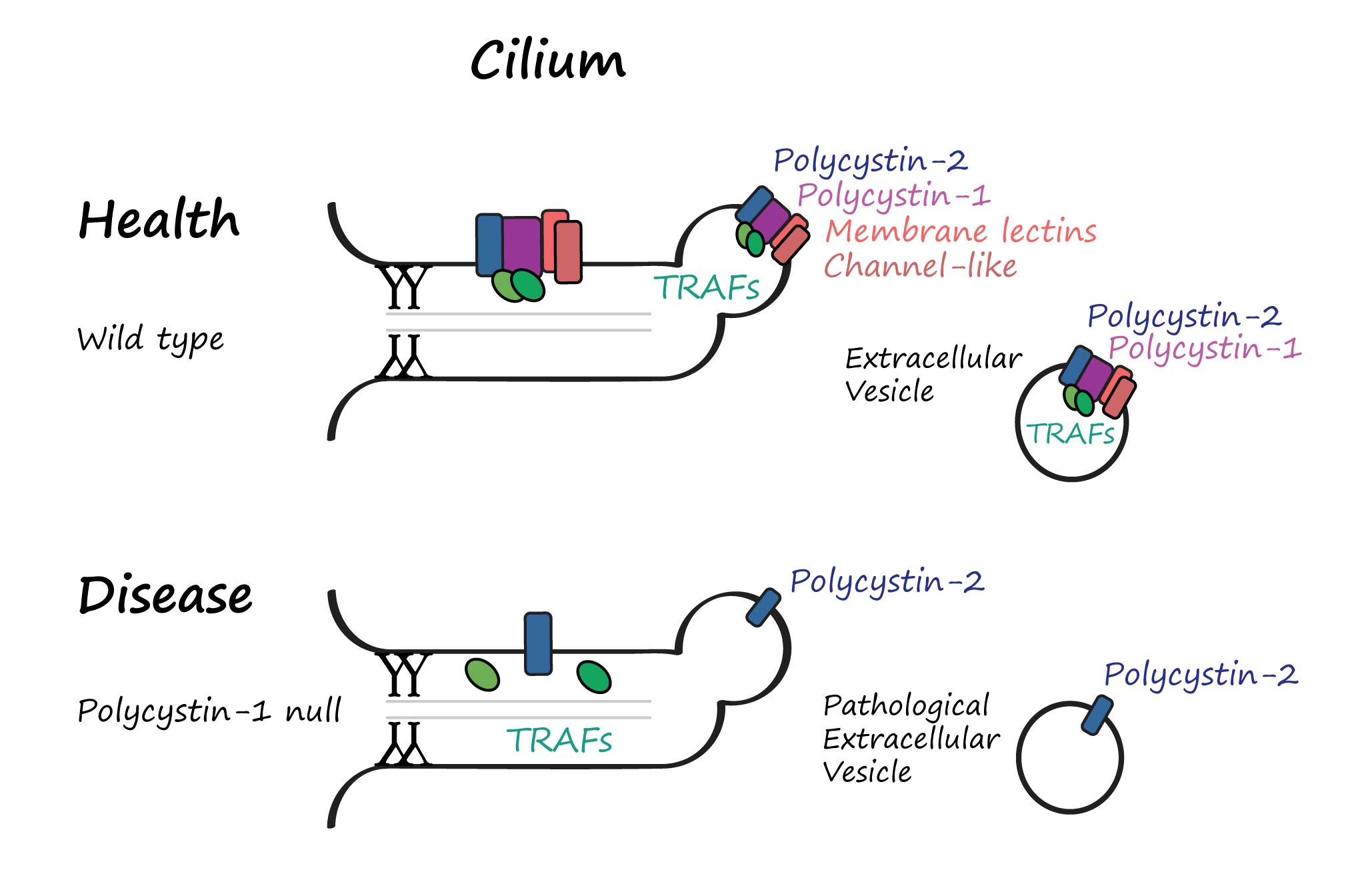
A Ciliopathy or an EVopathy?
We’re starting to think that what we call “ciliopathies”, such as polycystic kidney disease, may, in fact, be “EVopathies”—diseases where dysfunctional EV signaling, not just structural defects in the cilium, drives pathology. It’s a provocative hypothesis with far-reaching implications for diagnostics and therapies.
The beauty of model organisms is that they make the impossible possible. They let us decode the molecular script that underlies human biology.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Beautifully described Inna! Your talents as a scientist, artist and poet are all on display in this lovely primer about your model and the real world impact of your results. Well done, and congratulations!