UKB-MDRMF: A Framework for Multi-Disease Risk Prediction and Multimorbidity Assessment Using UK Biobank Data
Published in Healthcare & Nursing and Public Health

Overview of UKB-MDRMF
UKB-MDRMF is designed to transcend the limitations of traditional approaches by integrating multimorbidity mechanisms into disease risk prediction models.
Unlike methods focusing solely on individual diseases, UKB-MDRMF provides superior insights into disease-disease interactions, shared risk factors, and long-term health outcomes, offering a broader, more holistic understanding of human health trajectories.
Why UKB-MDRMF?
Challenges Addressed
-
Narrow Focus: Traditional models often miss cross-disease connections critical for comprehensive health management.
-
Fragmented Workflows: Disjointed processes limit the ability to perform integrated risk analysis.
-
Data Complexity: Combining diverse biomedical, lifestyle, genetic, and environmental data remains a major challenge in building robust predictive models.
Our Solution
By harmonizing multimodal data from the UK Biobank, UKB-MDRMF offers a powerful solution that improves predictive accuracy across a wide array of diseases.
Notably, 95.2% of disease categories demonstrated enhanced risk assessment performance when applying our framework, redefining standards in disease risk assessment.
How It Works
Step 1: Data Integration and Preprocessing
-
Multimodal Sources: We incorporated diverse data types including demographic information, lifestyle habits, physical measurements, environmental factors, genetic profiles (e.g., polygenic risk scores), and imaging data (brain and heart MRIs).
-
Variable Hierarchy: Variables were categorized into essential, detailed, and minor groups to optimize model relevance.
-
Rigorous Cleaning: We consolidated inpatient records, self-reports, and primary care data, partitioning the dataset into training, validation, and test sets with an 8:1:1 split to ensure robust, independent evaluations.
Step 2: Model Construction and Prediction
-
Prediction Models: We deployed a range of algorithms including Logistic Regression, Random Forest, XGBoost, and Fully Connected Neural Networks (FCNN).
-
Risk Assessment Models: We integrated time-to-event models such as Cox Proportional Hazards (CoxPH), DeepSurv, POPDxSurv, and CATISurv.
-
Joint Prediction: By simultaneously predicting multiple diseases (Phecodes), UKB-MDRMF captures both shared risk patterns and underlying multimorbidity mechanisms.
Step 3: Applications
-
Multimorbidity Discovery: Identification of latent relationships between diseases to inform prevention strategies and clinical decision-making.
-
Risk Factor Analysis: Quantitative evaluation of the contributions of lifestyle, environmental, and genetic factors to disease development.
-
Baseline Disease Risk Profiles: Establishment of comprehensive risk baselines across 1,560 diseases for future research and clinical use.
Conclusion
By integrating multimodal data and focusing on the interconnectedness of diseases, UKB-MDRMF presents a paradigm shift in how we approach disease prediction and prevention.
Its application spans from individual health management to large-scale public health strategies, marking an important step towards personalized, proactive healthcare.
Our publication in Nature Communications highlights the importance of developing scalable, integrative tools like UKB-MDRMF to tackle the complex reality of human health in a data-driven era.
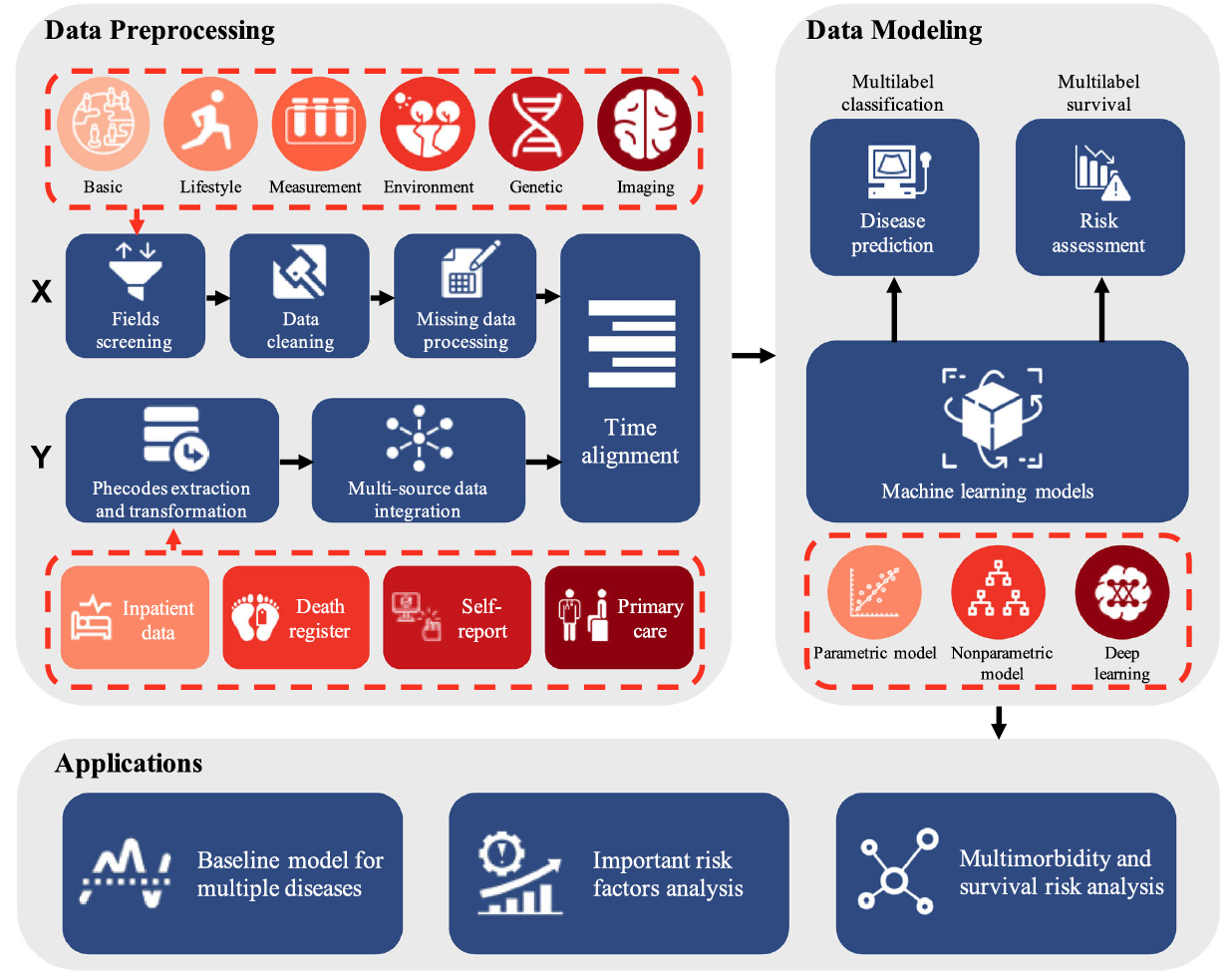
This pipeline utilizes input data from the diverse UK Biobank data, including six categories: basic, lifestyle, measurement, environment, genetic, and imaging data. Following field selection, data cleaning, and missing data preprocessing, predictors are generated. Response variables are derived from inpatient, self-reported, and primary care data, initially standardized to ICD-10 codes before conversion to Phecodes. After the temporal alignment of independent and dependent variables, the data is used to construct the UKB-MDRMF framework, encompassing disease prediction and risk assessment models. These models facilitate diverse applications, including establishing baseline conditions for multiple diseases, analyzing significant risk factors, exploring multimorbidity, and assessing survival risks. Icons are provided by Icons8 (https:// icons8.com).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in