What If We Could Study Membrane Proteins Without Membranes?
Published in Chemistry, Neuroscience, and Cell & Molecular Biology
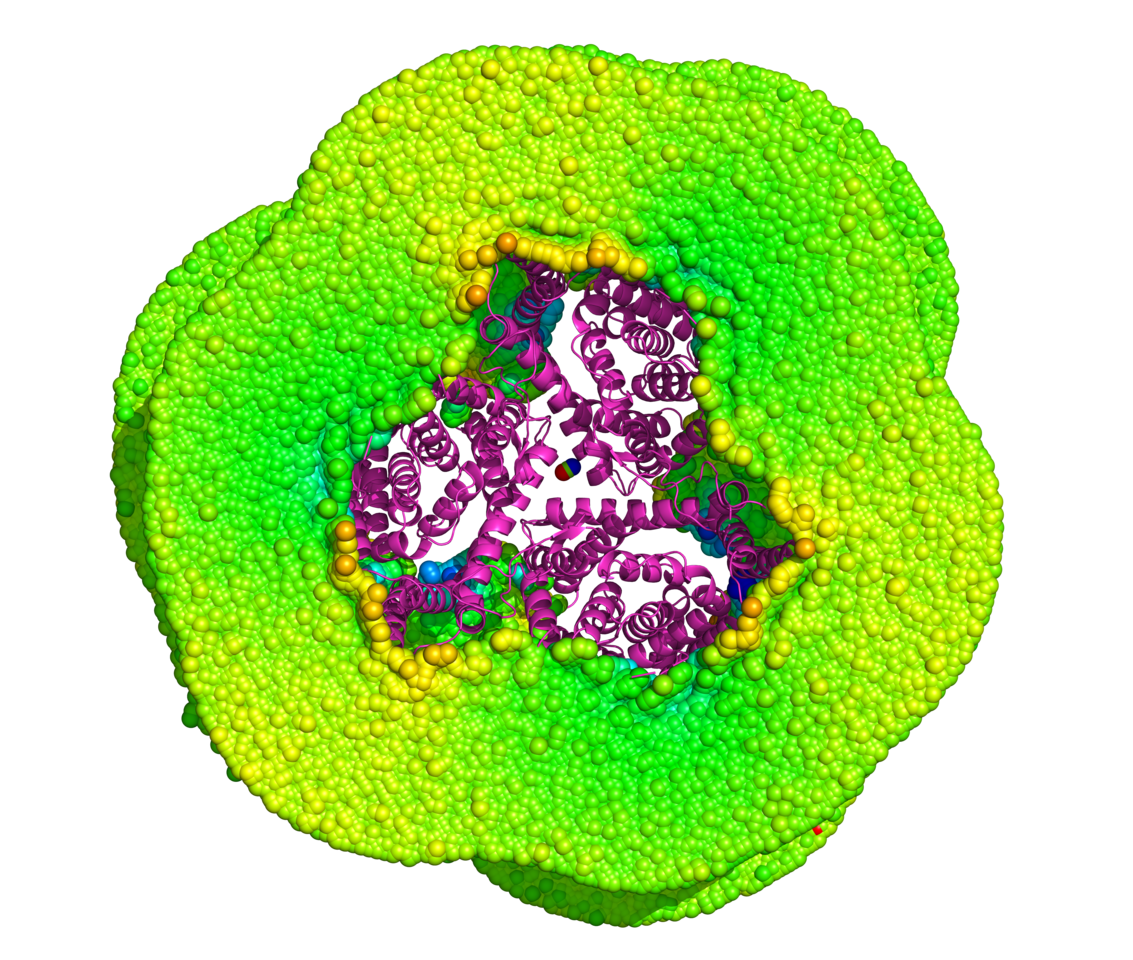
Every discovery begins with a moment of frustration.
Membrane proteins, especially transporters like EAA1, EAA2, and EAA3, sit at the heart of neuroscience, yet they remain frustratingly hard to study due to their insolubility and dependence on lipid bilayers. Every time experimentalists approached these proteins, they were forced to rely on detergents, nanodiscs, or membrane mimetics. But what if we didn’t need membranes at all?
Could a water-soluble mimic provide the same conformational insight as native membrane transporters embedded in lipids? This “heretical” question sparked the birth of our study.
From QTY Code to a Conceptual Experiment
The QTY code, developed to convert hydrophobic helices into hydrophilic ones while preserving 3D architecture, had already proven itself for GPCRs and chemokine receptors. But these were largely static evaluations: structure predicted, RMSD validated, case closed. We wanted to push it further.
-
Could a QTY-variant not only look like the native transporter, but actually move like it?
Could water interactions substitute for lipid head interactions at a dynamic level? - Would residue-wise fluctuations reflect evolutionary conservation of flexibility?
This was the origin of the molecular dynamics experiment.
The QTY-variants didn't collapse. They moved with a rhythm eerily similar to their lipid-embedded counterparts. Even their transition tendencies toward inward-facing states mimicked those seen in cryo-EM models.
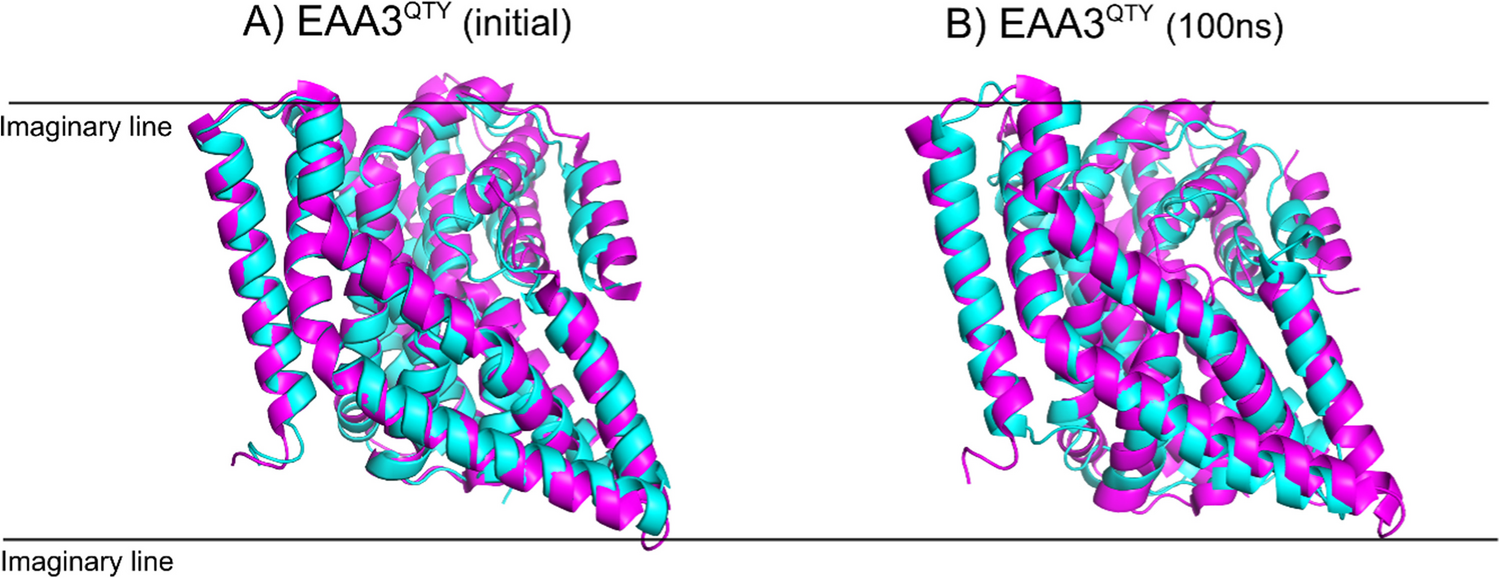
Conformational changes of water-soluble EAA3QTY and experimental outward and intermediate states of native EAA3. Superimposition of EAA3QTY structures from the MD simulation at 100 ns with experimental outward (8CV2) and intermediate (8CV3) states of EAA3. The RMSD values indicate significant structural alignment between the MD-simulated EAA3 and the experimental outward (1.137 Å) and intermediate states (2.347 Å), suggesting consistent conformational behavior. MD simulations were conducted for solutions, with Monte-Carlo placed K+ CL−ions (neutralizing, concentration = 0.15 M) https://doi.org/10.1007/s11095-024-03769-0
🎯 Did We Accidentally Build a Membrane Protein Evolution Model?
While analyzing residue-wise RMSF and RSA profiles, we noticed something unexpected regions with evolutionary conservation also coincided with dynamically conserved fluctuations in both systems.
This led us to a provocative speculation:
Have transporters evolved flexible cores that are intrinsically stable, independent of lipid presence with lipids acting as modulators rather than architects of dynamics?
If true, QTY-variants might represent an evolutionary abstraction of transporter dynamics, isolated from environmental bias.
Traditionally, QTY-engineering is framed as a pragmatic solubilization strategy. But our simulations invite a different interpretation. They expose dynamics that are lipid-independent. They reveal which motions are intrinsic vs. lipid-enforced. They hint that membrane proteins might be designed to fold and move correctly before encountering a membrane.
Follow the Topic
-
Pharmaceutical Research

Pharmaceutical Research is an official journal of the American Association of Pharmaceutical Scientists, covering innovative research in drug discovery, development, evaluation, and regulatory approval.
Your space to connect: The Psychedelics Hub
A new Communities’ space to connect, collaborate, and explore research on Psychotherapy, Clinical Psychology, and Neuroscience!
Continue reading announcement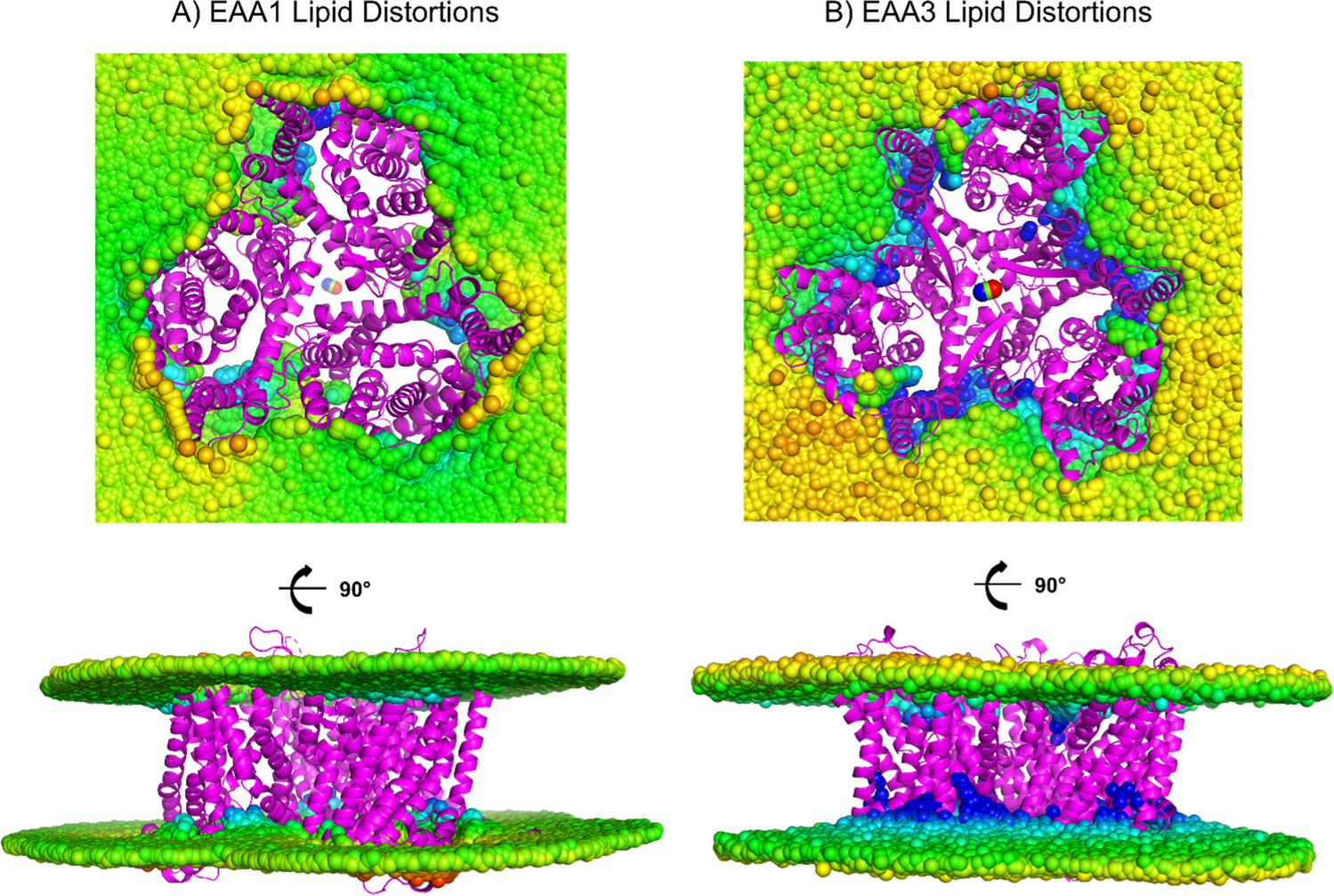

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in