When Half a Protein Is Enough
Published in Chemistry, Neuroscience, and Pharmacy & Pharmacology
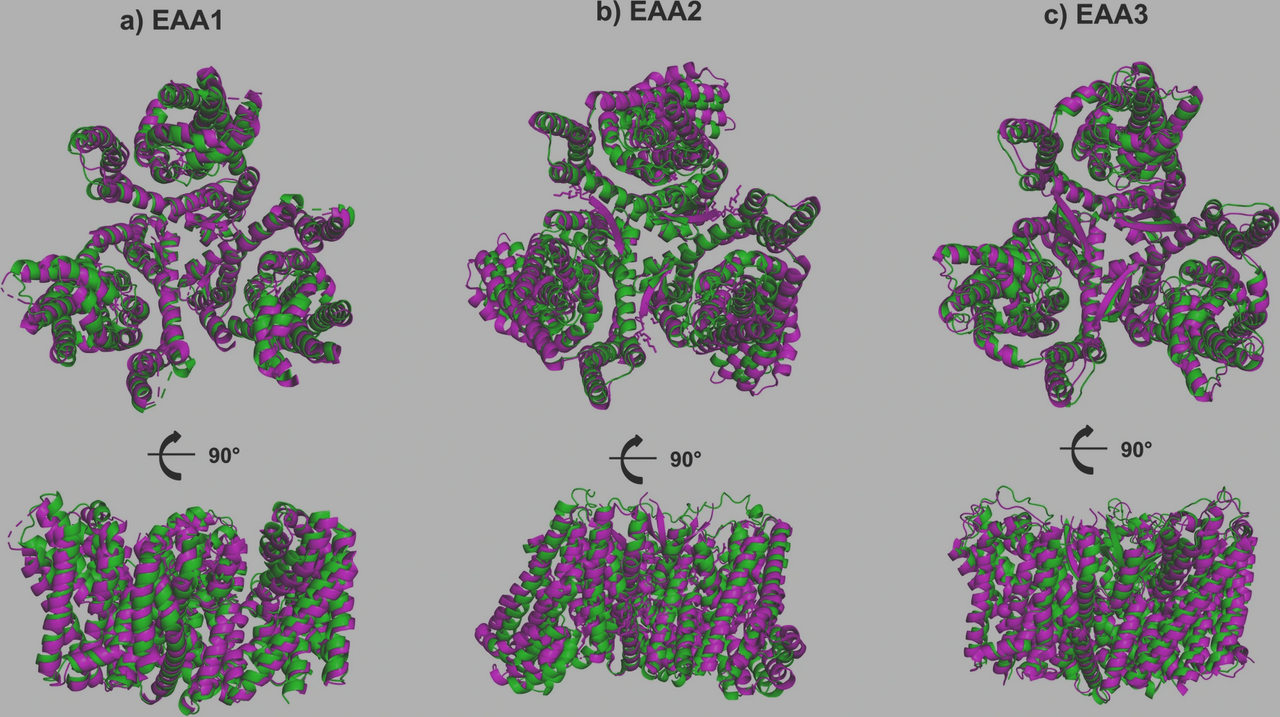
Glutamate transporters are essential gatekeepers of synaptic activity, preventing excitotoxic damage. For years, research focused on their canonical forms, full-length transporters that form trimers to maintain neurotransmitter balance. But biology is rarely so neat. Alternative splicing produces a multitude of truncated isoforms, often dismissed as incomplete or irrelevant. We too once viewed them this way. But while examining gene-centric isoform maps of glutamate transporters, we noticed something strange: many truncated isoforms still preserved entire transmembrane helices, especially those known to mediate oligomerization.
This raised a provocative question: If isoforms keep the structural fragments that allow transporters to form trimers, could they still interact with the full-length proteins?
At first, the idea sounded unlikely. After all, why would nature maintain incomplete proteins? But history offered clues. In G-protein coupled receptors (GPCRs), truncated variants had already been shown to act as negative regulators, binding their full-length partners and interfering with normal signaling (Qing et al. 2020, Li et al. 2023a , Li et al. 2023b). Could glutamate transporters harbor a similar hidden mechanism?
This “what if” moment set the stage. We started imagining truncated isoforms not as defective copies, but as molecular saboteurs, fragments that could slip into oligomerization sites and block proper assembly. If true, this would not only add an unexplored layer to synaptic regulation but also unveil new therapeutic opportunities, where isoforms (or their mimics) could inspire drugs that selectively dampen transporter activity in disease.
Using a combination of AlphaFold Multimer, docking pipelines, and molecular dynamics simulations, we placed truncated isoforms alongside canonical transporters in silico. To our surprise, several isoforms bound tightly to canonical assemblies, competing for the same trimerization interfaces. Notably, isoforms mimicking the TM2 and TM5 helices emerged as strong candidates for inhibitory interactions. In contrast, some isoforms displayed a fascinating twist: they preferred self-assembly, suggesting a protective mechanism that may buffer against interference with canonical transporters.
What we found
-
Truncated isoforms can act like molecular inhibitors, competing with full-length transporters for oligomerization surfaces.
Isoforms containing conserved helices (TM2, TM4, TM5) were particularly effective at mimicking trimerization domains. Binding interfaces were dominated by charged residues (Arg, Asp, Glu). In EAA2, specific Asp residues appeared only in isoform–canonical complexes, not in canonical dimers, making them attractive drug design targets. These charged contacts suggest that modulation of transporter oligomerization might be achievable with small molecules that mimic or disrupt isoform binding.
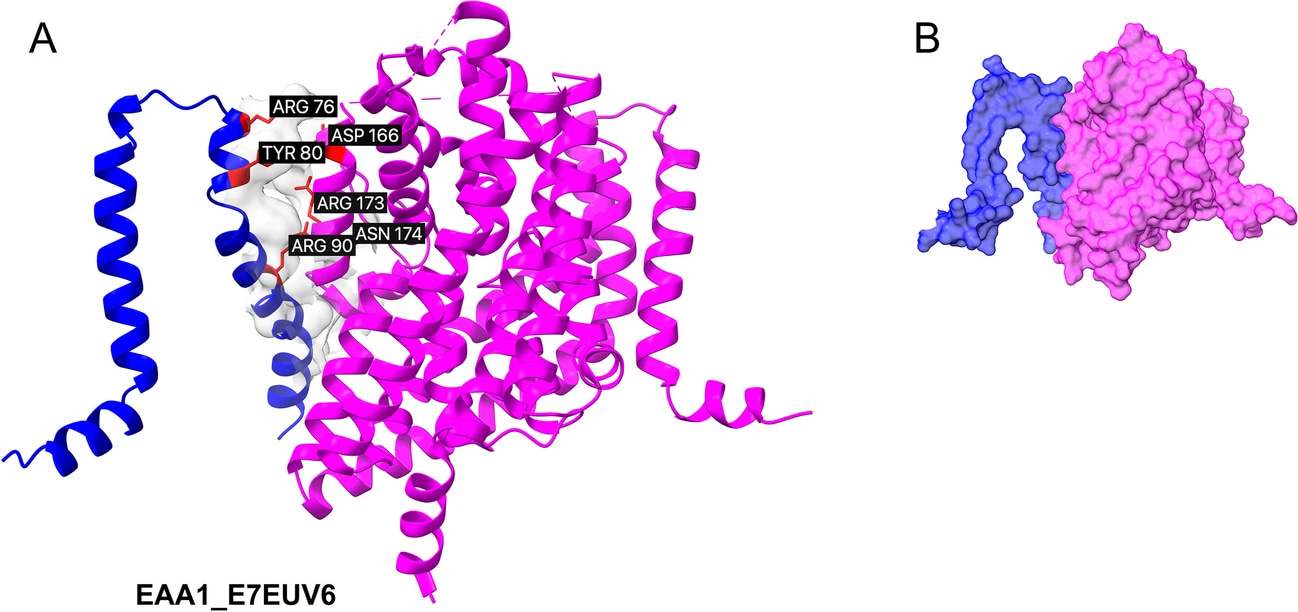
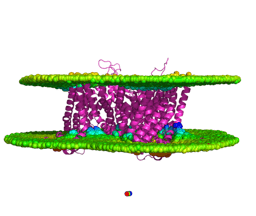
Heterodimeric complex containing canonical structure (EAA1) and the truncated isoform E7EUV6. Superposed canonical experimental transporter (magenta) and isoform structures (blue) are visualized. (A) The interface surface is displayed, with the six (three for the ligand and three for the receptor) highest energy-contributing residues highlighted in red and labeled. (B) The proposed competitive modulation of dimerization by the truncated isoform E7EUV6.
-
Evolutionary coupling analysis suggested these truncated helices are not random accidents, but preserved features with potential regulatory significance.
Why would evolution preserve fragments of transporters that appear incomplete? Our coevolution analysis provided an answer.
We found that the same helices that truncated isoforms preserve (TM2, TM4, TM5) are among the most conserved across glutamate, monoamine, and vesicular transporters. This suggests strong evolutionary pressure to maintain these domains, even in partial proteins. Even more intriguing: truncated isoforms showed residue co-evolution within intracellular regions, implying potential roles beyond inhibition, perhaps in signaling or scaffolding interactions with other proteins.
Not all isoforms were inhibitors.
Some, like A0A2R8Y4N0 (EAA2) and H0Y7R2 (EAA3), showed a remarkable preference for self-assembly, forming stable homodimers with binding energies as strong as −30 to −35 kcal/mol.
This hinted at a protective mechanism: instead of endlessly interfering with canonical transporters, certain isoforms may “soak up” their own disruptive potential by binding to themselves. In other words, nature may have evolved a way to limit collateral damage by allowing truncated isoforms to self-buffer.
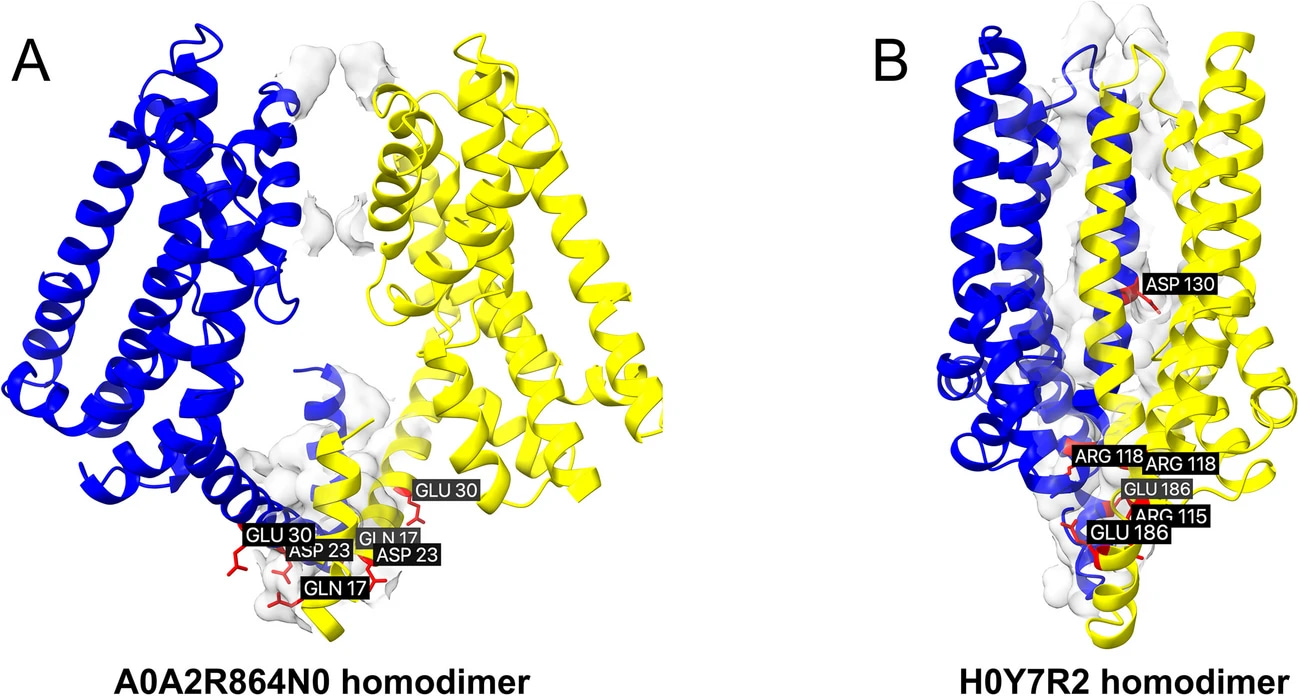
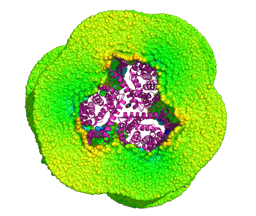
Self-assembly of A0A2R864N0 and H0Y7R2. Despite their lower ranking for canonical transporters, the isoforms are prone to self-assembly.
This work shifts the perspective on transporter biology. Instead of treating isoforms as passive byproducts, we highlight them as active modulators of protein–protein interactions. These results indicate that truncated isoforms could directly inhibit trimerization, acting as natural modulators of transporter efficiency. From a therapeutic standpoint, they could serve as natural templates for inhibitor design -guiding drugs that block pathological transporter activity in neurodegenerative or psychiatric diseases.
Follow the Topic
-
Pharmaceutical Research

Pharmaceutical Research is an official journal of the American Association of Pharmaceutical Scientists, covering innovative research in drug discovery, development, evaluation, and regulatory approval.
Your space to connect: The Psychedelics Hub
A new Communities’ space to connect, collaborate, and explore research on Psychotherapy, Clinical Psychology, and Neuroscience!
Continue reading announcement
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in