When defense systems facilitate the emergence of antibiotic resistant lineages
Published in Microbiology
Antibiotic resistance is of major public health concern, having been responsible for more than 1.2 million deaths in 2019 1. In particular, multiresistant and even panresistant bacterial lineages are emerging and disseminating, resulting in therapeutic failures. Carbapenems are one of the last resort antibiotics used to treat infections caused by multidrug-resistant (MDR) Gram-negative bacteria. However, the number of Carbapenemase-producing Enterobacteriaceae, resistant to these drugs, is on worldwide rise. Among them, Carbapenemase-producing Escherichia coli (CP-Ec) are particularly feared as these bacteria are common inhabitant of the intestinal microbiota able to spread in the community. Dissemination of genes encoding carbapenemases in Enterobacteriaceae is mainly linked to horizontal transfer of plasmids. These plasmids differ by their transmission efficiency through conjugation and by the metabolic burden they induce in their bacterial host.
We previously characterized the diversity and evolution of CP-Ec isolated in France before 2016, and showed that carbapenemase genes are associated with a wide variety of E. coli genomic backgrounds and a small number of dominant phylogenetic lineages2. The most frequent carbapenemase gene was blaOXA-48, encoding the OXA-48 carbapenemase, reflecting a general tendency in Europe. This is attributed to the high conjugation rate of the IncL pOXA-48 plasmids. However, in Sequence Type ST38 isolates, the blaOXA-48 is often carried on the chromosome rather than on plasmids3. ST38 isolates belong to Extra-Intestinal Pathogenic E. coli (ExPEC) and represented the most abundant lineage in our study. By using a phylogenetic approach, we showed that four ST38 sublineages, characterized by different blaOXA-48chromosomal insertions, were disseminating in France, and for three of them worldwide2. Intriguingly, while these disseminating isolates were multiresistant, they carried only few if any plasmids.
Antibiotic resistance gene (ARG) chromosomal integration is considered as a product of plasmid long-term evolution, mainly under selective pressure. Whether any factors are required to increase the frequency of these integrations in some bacterial lineages was unknown.
In our paper published in Nature Communications, we used the E. coli ST38 lineage as a model to explore this question. We have developed an experimental evolution strategy to reproduce blaOXA-48 integration into the chromosome. We transferred into three different ST38 isolates devoid of carbepenemase genes, different pOXA-48 plasmids. Plasmids and strains were selected based on our evolutionary data on ST38. We observed that, in the transconjugants, the pOXA-48 plasmids induce different fitness costs and have low stability in the absence of antibiotic selection (Figure. 1).
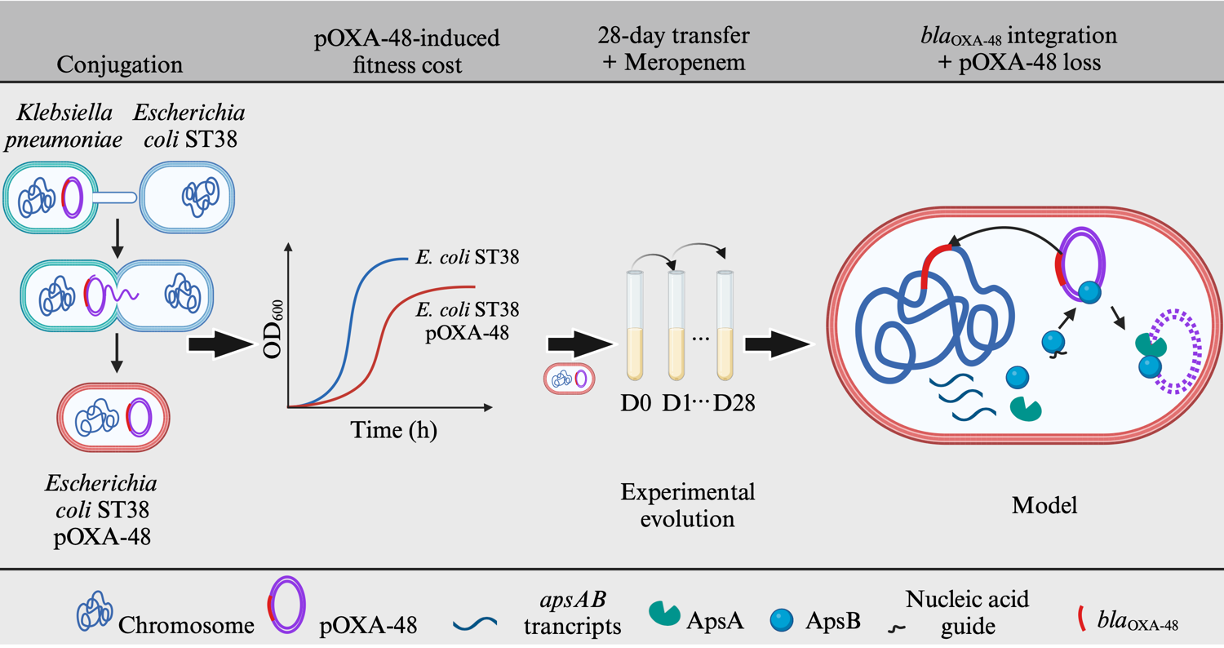
Figure 1: The journey of E. coli ST38/pOXA-48 transconjugant to emergence of blaOXA-48 chromosomally integrated lineages following experimental evolution.
We performed a 28-day evolution experiment in the presence of a subinhibitory concentration of meropenem and parallelly monitoring the evolution of the fitness of the whole population. We observed a rapid fitness improvement in a specific ST38/pOXA-48 combination during the experimental evolution. DNA sequencing of the pooled population showed two main populations from this evolved combination: a population 1, representing more than 99% of the whole population, that had lost the plasmid but carried blaOXA-48 in a new replicon and a minor population 2 still carrying the pOXA-48 plasmid. By analyzing genetic changes arisen in this evolved combination, we found that in population 1 blaOXA-48 is rapidly integrated into the chromosome or the resident F plasmid and pOXA-48 plasmid lost. Conversely, population 2 kept the plasmids but was preferentially mutated in a two gene operon (ApsAB). We next combined evolutionary experiments using different strains and plasmid to show that integration of blaOXA-48 in a new replicon depends on three main factors (Figure. 1): 1) the presence of the resistance gene in an active mobile genetic element, such as a transposon; 2) a high plasmid-induced fitness cost on the host and 3) an active elimination of the pOXA-48 plasmid by ApsAB. Indeed, the analysis of the phenomenon of ARG integration, led us to the discovery of a novel antiplasmid system. In this system, ApsB is an Argonaute-like protein that lacks the endonuclease activity but is potentially able to bind small nucleic acid guides that direct plasmid recognition. ApsB was proposed to interact with ApsA that contains a helicase and a nuclease domains and is likely responsible for plasmid degradation. ApsAB can destabilize low and high copy number plasmids. We identified related systems in various bacteria phyla from Enterobacteriaceae to cyanobacteria and proposed that they have influenced plasmid dissemination and antibiotic resistance gene integration in those phyla too.
References
- Murray, C. J. L. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399, 629–655 (2022).
- Patiño-Navarrete, R. et al. Specificities and Commonalities of Carbapenemase-Producing Escherichia coli Isolated in France from 2012 to 2015. mSystems 7, e01169-21 (2022).
- Turton, J. F. et al. Clonal expansion of Escherichia coli ST38 carrying a chromosomally integrated OXA-48 carbapenemase gene. J. Med. Microbiol. 65, 538–546 (2016).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in