A cancer rainbow mouse for visualizing the functional genomics of oncogenic clonal expansion
Published in Cancer
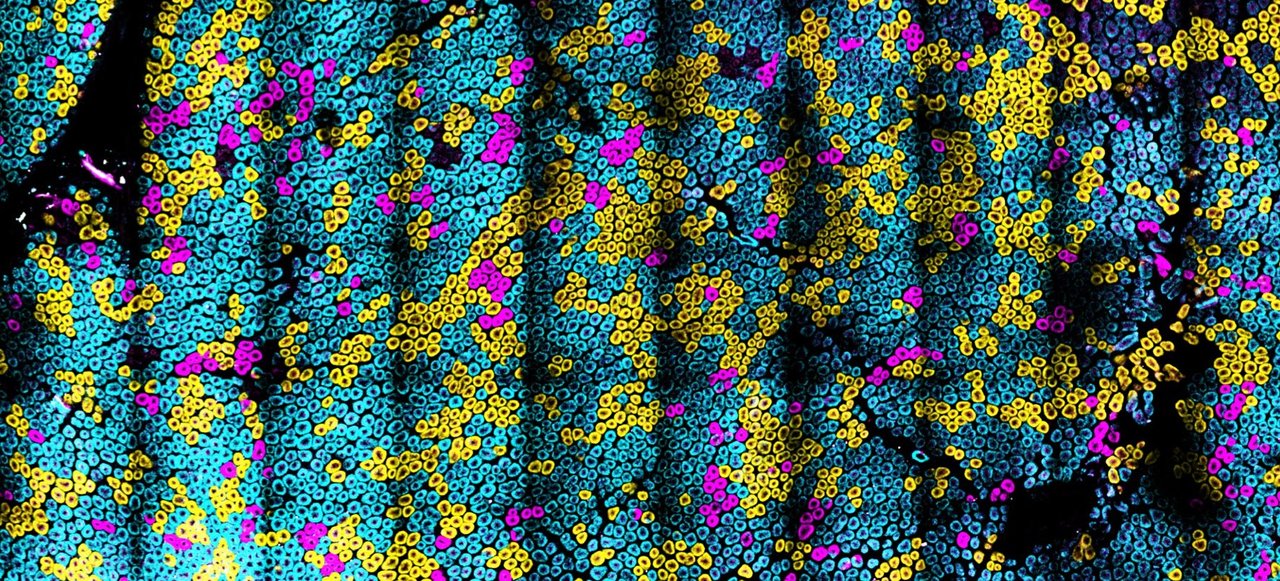
Preventing the spread of somatic mutations in the epithelium could provide new strategies for reducing the risk of cancer. Therefore, our goal was to develop genetically tractable model systems for visualizing this process. A clue for how to proceed came during my postdoctoral fellowship. As a post doc, I needed to compare the effects of multiple receptor isoforms in vivo. However, modeling these receptors and comparing their activity in vivo was fraught with many challenges, most notably the need to make mouse lines for each receptor of interest. Not only was this a financial roadblock, but a scientific one as well – comparing multiple transgenic mouse lines will always be labor intensive and a source of significant experimental error. We realized that one answer to this problem was to encode multiple receptor isoforms with a fluorescent protein for barcoding. In this way we could use lineage tracing to report on the effects of each receptor at the cellular level – similar to the Brainbow approach1 .
Our approach successfully enabled us to study LGR5 mutations in vivo2, but several limitations were encountered. First, the signal from the fluorescent proteins was unreliable and required antibody staining to boost their signal above background. Second, the genetic engineering and cloning was arduous. In the current paper, my lab uses a new approach to interchangeably shuttle oncogenes into Crainbow based imaging vectors that can be easily transferred into mice. In our paper, we tested and identified optimal fluorescent proteins for expression in vivo – with no antibody staining – that also survived fixation and permitted live imaging. Each fluorescent protein was also spectrally resolvable and adaptable for mRNA FISH and histopathology coregistry. We also developed staining strategies for subcellular localization of each oncogene. In short, our Cancer rainbow (Crainbow) model provides a plug-and-play approach for expressing and visualizing the effects of multiple oncogenes in the same mouse with spatiotemporal precision.
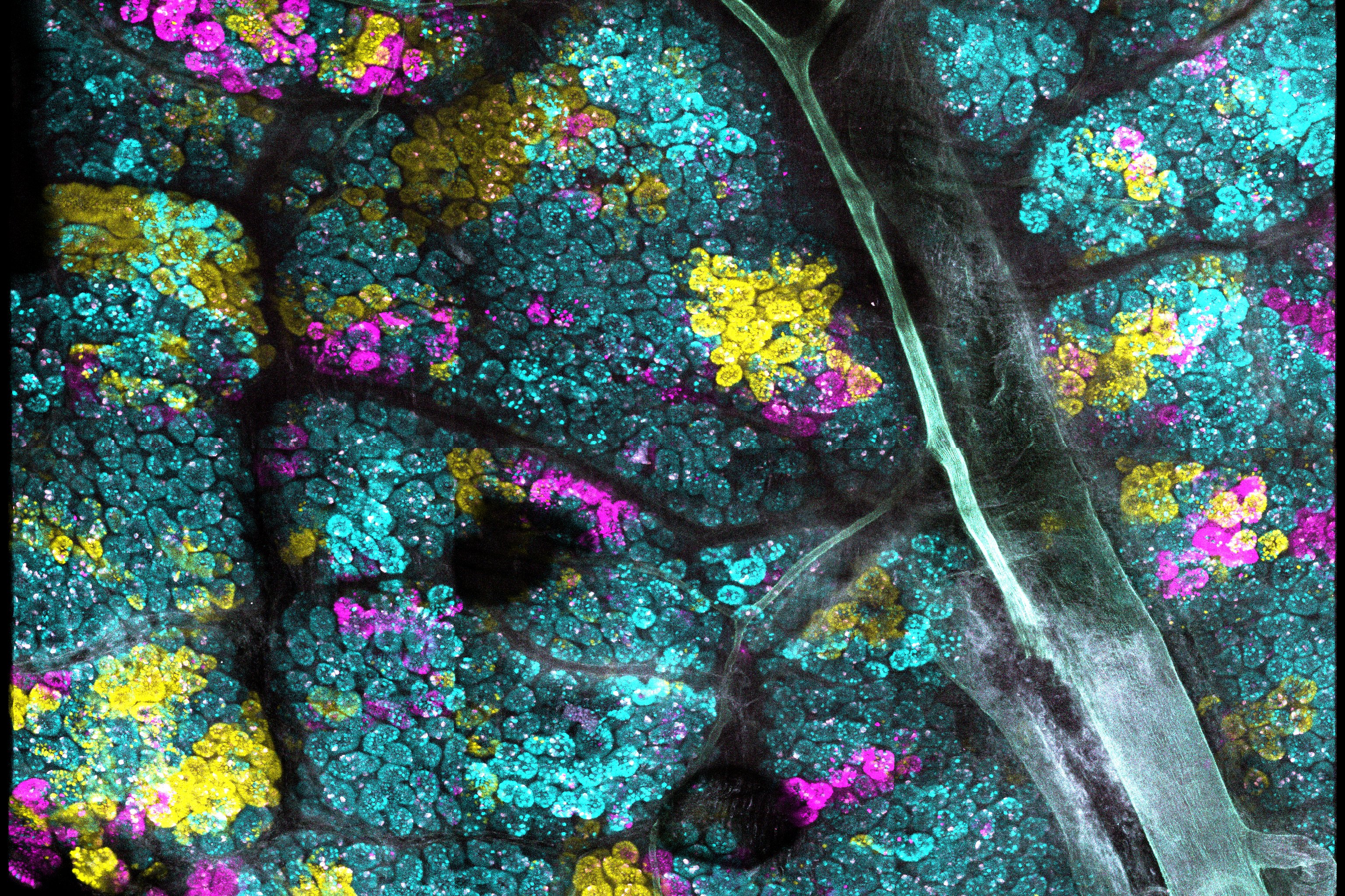
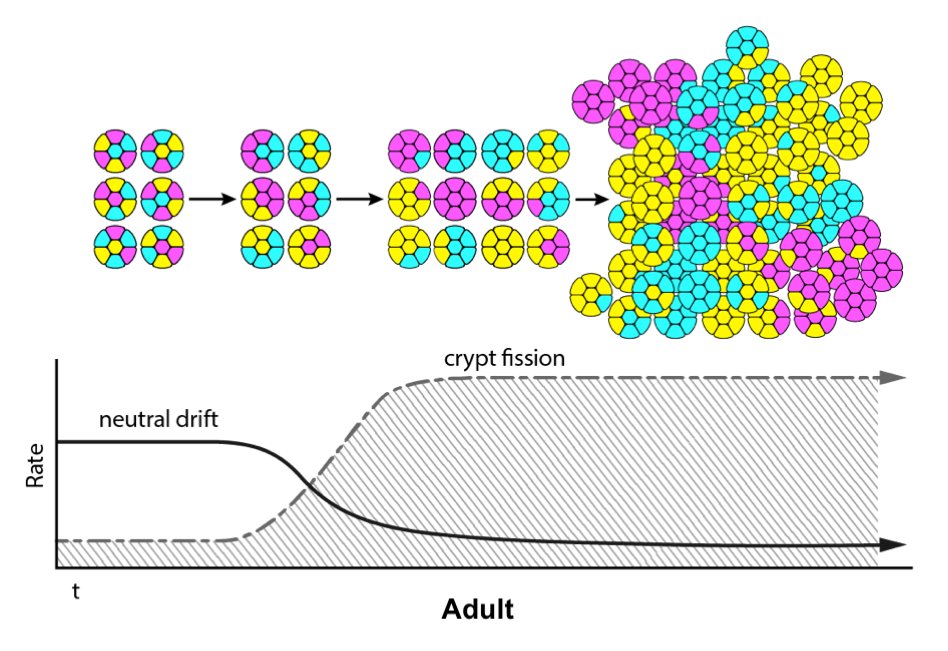
Crainbow mouse models could be useful in many ways – one could study tumor heterogeneity, visualize metastasis, or compare oncogenic potential. We decided in this first paper to use Crainbow for studying how somatic mutations acquired in the intestinal stem cell spread throughout the epithelium. We initially chose the intestine for our work – other organs and cancers are being studied currently – because of the well-documented activity of intestinal stem cells residing at the crypt base. Crainbow was used to empirically show that the lateral spread (i.e. the spread of mutations from one crypt stem cell into neighboring crypts) was very inefficient. Even potent oncogenes, like ßcatenin, were unable to significantly spread laterally in adults. A surprising finding was that somatic mutations rapidly spread during postnatal development –a critical period in which crypts are more dynamic than in adults. This finding suggests that colorectal carcinogenesis may be seeded during the first few years of life. Lastly, we found a new functional role for the oncogenic fusions of Rspondin-3 as drivers of somatic spreading – even in adults. This appears to occur by increasing crypt fission and reducing the neutral drift behavior of intestinal crypts, a process we term “microenvironmental oncogenesis” because it extrinsically modifies stem cell behavior.
Moving forward, we intend to increase the repertoire of validated Crainbow models to study the spread of somatic mutations and visualize the progression of precancers to invasive cancers. Read our full paper here: https://www.nature.com/articles/s41467-019-13330-y
1 Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56-62, doi:http://www.nature.com/nature/journal/v450/n7166/suppinfo/nature06293_S1.html (2007).
2 Snyder, J. C. et al. Inhibiting clathrin-mediated endocytosis of the leucine-rich G protein-coupled Receptor-5 diminishes cell fitness. J Biol Chem, doi:10.1074/jbc.M116.756635 (2017).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in