A New Approach to High Accuracy Nanopore Sequencing
Published in Bioengineering & Biotechnology
Nanopore DNA sequencing is a single-molecule DNA sequencing technology that has over the past decade matured from an idea, into a proof-of-concept, and finally into a working sequencing technology. Nanopore sequencing has gained attention for its considerable advantages, including long reads and direct detection of epigenetic markers. However, nanopore sequencing has also become known for its main remaining limitation: its low single-read sequencing accuracy relative to other technologies. While much work has been done to improve base calling algorithms, we attack the low accuracy problem from another direction by re-engineering the nanopore signal to produce more information from a read.
Traditional nanopore sequencing applies a constant voltage to drive DNA through a nanopore while the ion current through the pore is measured to identify the bases. A motor enzyme is used to control the DNA’s motion through the pore in discrete steps (Fig 1a). The key insight of our work is that nanopore sequencing can access more information about the translocating DNA by probing the DNA at positions in between the discrete steps that the motor enzyme provides. When the constant transmembrane voltage is replaced with a time-varying voltage, the DNA within the pore is stretched according the voltage. Thereby, the voltage provides a fine control over the DNA position between the coarse step-control provided by the enzyme (Fig 1b). Using this trick, we measure the DNA at all positions along its length rather than only at discrete positions, thus generating a richer signal that we can use to combat major sequencing error modes. Particularly, the new signal can be used to infer motor enzyme missteps that confound base calling algorithms. Furthermore, this method distinguishes DNA sections which produce indistinguishable signatures in constant voltage operation. While constant voltage produces reads barely better than random base calling, the variable voltage method produces base call accuracy near 80% with just one single-stranded DNA read.
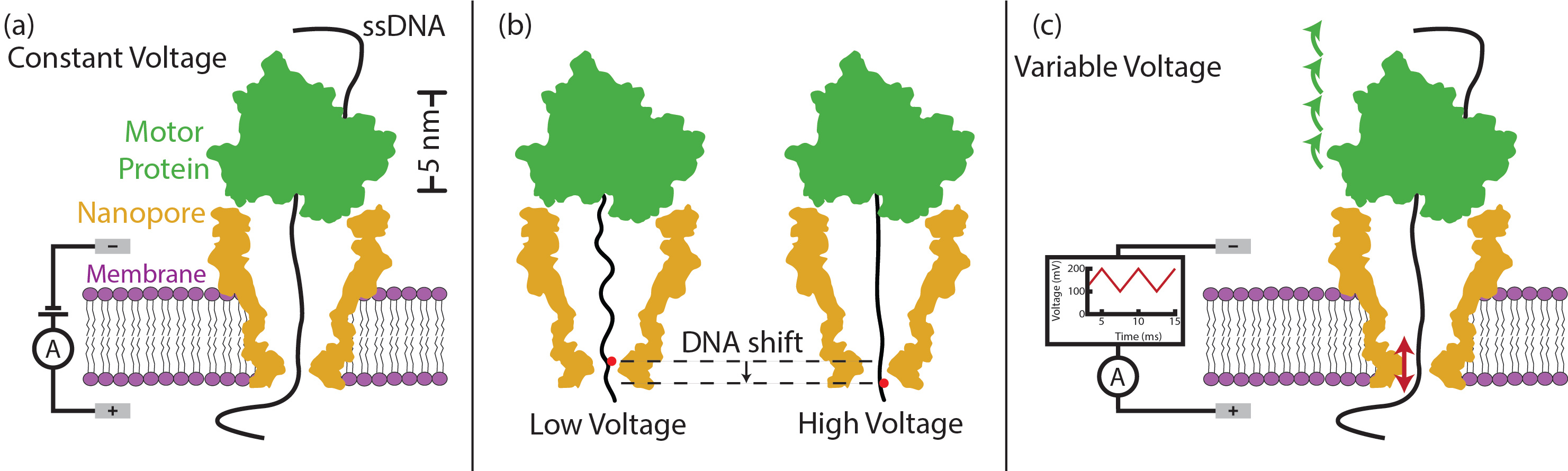
Figure 1 (a) Regular nanopore sequencing applies a constant voltage across the membrane (purple), driving single-stranded DNA (black) through a nanometer-scale pore (gold). A motor enzyme (green) controls DNA motion through the pore. (b) Applying different voltages changes the force pulling on the DNA. Higher forces cause the DNA to extend further, shifting the DNA within the constriction of the pore and changing the bases sensed by the nanopore. (c) In variable voltage sequencing, we vary the voltage at 200 Hz, repositioning the DNA within the pore (red arrows) while the motor enzyme steps along the DNA at an average rate of ~20 Hz (green arrows).
An exciting aspect of the new nanopore operation method is that it maintains all the advantages of nanopore sequencing, such as long reads and speed, and it is complementary to most other efforts that improve sequencing accuracy. For example, it can be used in conjunction with new machine learning methods and base calling algorithms, as well as improved pores pushing nanopore sequencing accuracy even higher. The variable voltage method may be adapted into existing nanopore sequencing platforms enabling another leap forward in accuracy.
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in