Beyond single gene association studies: the need for new approaches to explore the complexities of chlamydiosis in koalas.
Published in Genetics & Genomics and Zoology & Veterinary Science

Host pathogen interactions are complex, dynamic and often poorly understood in species of conservation concern. Understanding host genetic factors contributing to pathogenesis, including susceptibility and resistance, is crucial in preserving threatened species. The koala, an iconic Australian marsupial, is facing dramatic population declines resulting in some populations being listed as “Endangered”, with continued environmental and pathogenic challenges. Chlamydiosis, caused by the bacterium Chlamydia pecorum, is one of the most common infectious diseases impacting koalas, severely impacting health and fertility. The bacterium infects the mucosal cells of the ocular and urogenital tracts, causing inflammation and fibrosis of infection sites, often leading to para-ovarian cysts and infertility. Many host, pathogen and environmental factors have been linked to chlamydiosis outcomes, however host genetics has been a recent focus of disease studies.
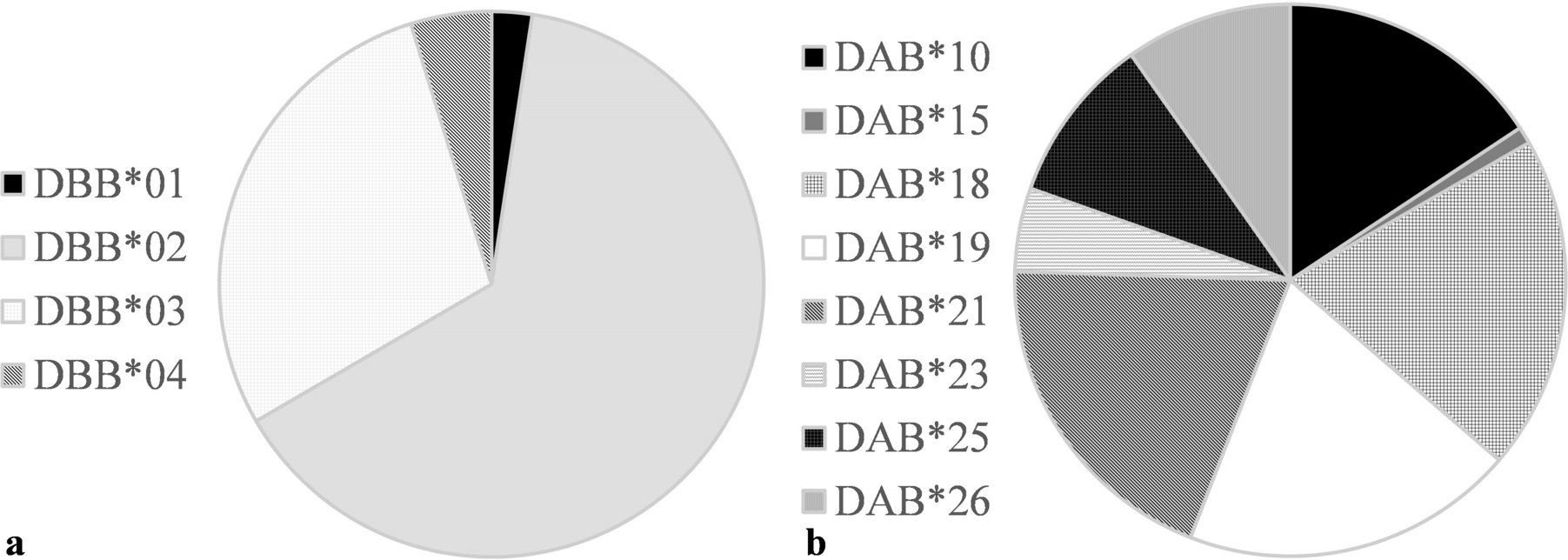
The Major Histocompatibility Complex (MHC) is a key component of the immune response, responsible for the presentation of antigens to CD4 T-cells. MHC diversity is assumed to influence immunological responses, through an increased repertoire of peptide binding regions. It is hypothesized that the increased diversity of DA and DB beta subfamilies of MHCII reflects selection pressures in koalas. While MHCII has been associated with chlamydiosis in several species, including koalas, these findings differ between studies and the role of MHC remains unclear. It is possible that differences in sampling, experimental and analysis methodologies represented in the literature as well as the complexity of disease definition in studies on wildlife may have caused these ambiguous results.
During this study we utilized high-throughput amplicon deep-sequencing of wild koalas from the Gunnedah region of New South Wales, to explore MHCII DAB and DBB gene variants. Koalas within the Gunnedah region have been extensively studied for the past few decades and have shown a notable increase in chlamydiosis prevalence and severe clinical signs in the last 10 years, alongside a dramatic decline in population numbers, providing a unique opportunity to explore disease under these harsh natural conditions. It was observed that many females in Gunnedah had developed para-ovarian cysts resulting from chlamydiosis, a potential cause of population decline through infertility reducing the breeding population. These “infertile” female koalas were compared to “fertile” female koalas from the same location who continued to carry young, to provide tightly defined disease and control groups. As this population had been studied longitudinally, we were able to compare findings from a similar study1 on the same population collected 10-years previous, as well as comparing a group of juvenile koalas to the adult cohort, to explore generational changes and potential disease selection pressures at the MHCII loci.
Key Findings
Generational selection of increased MHC variants per individual may have been detected in this population, supporting previous hypothesis of advantages of increased diversity at MHC. When comparing findings from a previous study1, there was a signficant increase in mean number of DAB alleles per individual in the current study and a signficant difference in allele frequency of DAB. However, the opposite was also observed when comparing age cohorts, juvenile koalas had significantly decreased number of DBB alleles compared with adults. In both contexts the cohorts with increased mean number of MHC variants were those with greater sample size, so may be a result of sampling bias or chance rather than generational selection.
MHCII DAB may have an impact on chlamydiosis-induced infertility, supporting previous research. The allele frequency of the DAB loci was significantly different between infertile and fertile disease groups, however individual alleles contributing to this difference could not be identified possibly due to the small sample size resulting from the strict inclusion criteria of disease definitions.
Challenges
The MHC region is notably complex and highly variable among individuals. Some MHC genes often appear in multiple copies making it difficult to assign alleles to corresponding loci, creating barriers to drawing definitive conclusions. This is where long read sequencing and whole genome sequencing can be utilized to reveal the true variation of these intricate regions.
In the context of wildlife studies, another challenge faced is the limited access to samples and the inherent uncertainties of natural diseases. The use of tightly defined disease groups did not resolve the ambiguity of disease definitions observed in previous research and further limited our sample sizes. A solution to such challenges could be the use of longitudinal monitoring of several koala populations across their range, coupled with consistent data collection, providing more reliable comparative groups for genetics research.
Future Directions
The findings from this study suggest that while MHCII variants likely have a role in koala immune responses to chlamydiosis, uncovering such complex pathogenesis requires a shift from single gene association studies to more holistic approaches. Future studies should consider the utilization of multifactorial approaches, such as genome-wide association studies utilizing longitudinal data from multiple populations, to provide a comprehensive understanding of the multiple genetic pathways involved in complex immunological interactions and underlying chlamydiosis outcomes in koalas. This study highlights the complex nature of pathogenesis in wildlife contexts and provides a guide for future investigations of host genetic involvement in koala chlamydiosis, to inform targeted koala conservation across its diverse range and host-pathogen-environment contexts.
References
- Lau, Q., Jaratlerdsiri, W., Griffith, J. E., Gongora, J. & Higgins, D. P. MHC class II diversity of koala (Phascolarctos cinereus) populations across their range. Heredity (Edinb) 113, 287-296 (2014). https://doi.org:10.1038/hdy.2014.30
Follow the Topic
-
Scientific Reports

An open access journal publishing original research from across all areas of the natural sciences, psychology, medicine and engineering.
Related Collections
With Collections, you can get published faster and increase your visibility.
Obesity
Publishing Model: Hybrid
Deadline: Apr 24, 2026
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in