Seeing the Unseen: Antibiotic-Derived Fluorescent Probes to Visualise Bacteria
Published in Microbiology

Antimicrobial resistance is a global concern that increasingly threatens modern medicine. Compounding the decline in antibiotic effectiveness is the retreat of most major pharmaceutical companies from antibiotic research and development, and the market failure of small companies attempting to fill the gap. New antibiotics are urgently needed, along with diagnostic tests capable of rapidly identifying resistant bacteria to improve antibiotic stewardship. In order to develop new drugs that circumvent existing resistance mechanisms, while themselves withstanding the rapid development of resistance, we must first improve our fundamental understanding of how antibiotics interact with, and kill, bacteria.
Over the last 10 years we have been developing a unique set of antibiotic-derived fluorescent probes to investigate how antibiotics interact with bacterial and human cells at a molecular level. We want to apply these tools to study the mechanistic basis of drug-bug interactions and innovate rapid resistance profiling assays.
Our approach is based on the derivatisation of known antibiotics by attaching a readily modified (azide) functional group at a site where structure-activity relationship (SAR) data sourced from existing literature indicates modifications are tolerated (Figure 1). The azide substituent provides a functionalised handle that allows a range of different colour alkyne-derivatised fluorophores to be easily attached for each antibiotic. Fluorophore conjugation is accomplished by facile coupling reaction using the highly selective “click” Cu(I)-catalysed alkyne−azide cycloaddition (CuAAC) coupling reaction. The resulting triazole linkage is chemically and biologically stable and non-reactive.
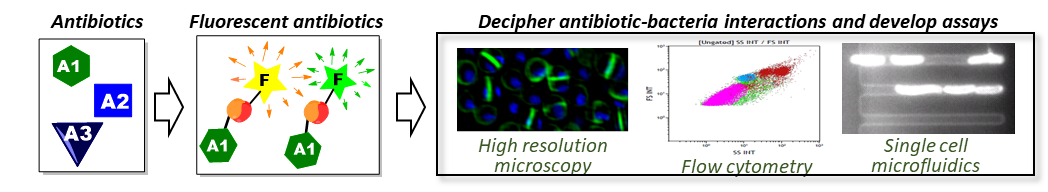
This research article describes the synthesis and application of a set of probes derived from the glycopeptide vancomycin, an antibiotic that works against Gram-positive bacteria by binding to Lipid II, a precursor of the cell-wall peptidoglycan. We selected vancomycin as we had extensive experience in preparing vancomycin analogues through a research project developing ‘supervancomycin’ therapeutic candidates [1,2], and knew that there was significant flexibility in what could be attached to vancomycin’s C-terminal end without affecting activity. While other vancomycin probes have been previously described, and indeed are commercially available, they have not been widely utilised, and they show some disadvantages when compared to our new probes. Importantly, our vancomycin probes retain a similar activity profile to the parent antibiotic, ensuring that they are valid surrogates for experiments examining how vancomycin works.
High resolution confocal microscopy studies using the vancomycin probes clearly showed binding to the peptidoglycan cell wall of Gram-positive bacteria such as Staphylococcus aureus, with a high concentration at the dividing septum, the site of vancomycin’s target Lipid II. An advantage of the click chemistry approach is the flexibility it allows to choose from multiple colour fluorophores, particularly beneficial when co-staining the bacteria with other dyes. As expected, the vancomycin-NBD probe did not normally stain Gram-negative bacteria such as Escherichia coli, since the outer-membrane prevents penetration of the vancomycin to reach the peptidoglycan layer. However, we saw that occasionally cells were labelled at the point of bacterial cell division. This prompted testing in a membrane-damaged mutant, which showed substantial internal labeling. This finding inspired development of an assay for outer membrane damage. Antibiotics not damaging the membrane gave no signal, while membrane-active antibiotics produced strong fluorescence. This mechanism-specific assay has already been shown to be of utility to collaborators [3], and we think it has potential to become widely used in assays assessing the potential mechanism(s) of action of new antibiotics.
Importantly, a key reason we are making these antibiotic-derived probes is to create research tools to facilitate the work of other research teams. We strongly believe that microbiologists around the world are much more likely to be able to imagine innovative applications of the probes than we are (particularly since we have a medicinal chemistry background!), so are keen to collaborate and share the probes In addition to the vancomycin derivatives included in this paper, we have previously published on probes derived from trimethoprim [4], linezolid [5], ciprofloxacin [6], and roxithromycin [7], with unpublished probes also available (or in progress) that are derived from polymyxin, octapeptin, tachyplesin-1, arenicin-3, daptomycin, tetracycline, neomycin, and a range of lactam antibiotics. Hopefully, together we can turn the tide against resistant superbugs!
References:
[1] Blaskovich et al. A Lipoglycopeptide Antibiotic for Gram-Positive Biofilm-Related Infections Science Translational Medicine 14, 662, eabj2381 (2022) DOI: 10.1126/scitranslmed.abj2381
[2] Blaskovich et al. Protein inspired antibiotics active against vancomycin and daptomycin resistant bacteria”; Nature Communications (2018) 9, 22 (2018). DOI:10.1038/s41467-017-02123-w.
[3] Oliveira et al. Rescuing Tetracycline Class Antibiotics for the Treatment of MDR A. baumannii Pulmonary Infection. mBio 13, e03517 (2022). DOI: 10.1128/mbio.03517-21
[4] Phetsang et al. Fluorescent Trimethoprim Conjugate Probes to Assess Drug Accumulation in Wild Type and Mutant E. coli. ACS Inf. Dis. 2, 688 (2016). DOI: 10.1021/acsinfecdis.6b00080
[5] Phetsang et al. An azido-oxazolidinone antibiotic for live bacterial cell imaging and generation of antibiotic variants. Bioorg. Med. Chem. 22, 4490 (2014). DOI: 10.1016/j.bmc.2014.05.054
[6] Stone et al. Fluoroquinolone-derived fluorescent probes for studies of bacterial penetration and efflux. MedChemComm 10, 901 (2019). DOI: 10.1039/C9MD00124G
[7] Stone et al. Fluorescent macrolide probes – synthesis and use in evaluation of bacterial resistance. RSC Chem. Biol. 1, 395 (2020). DOI: 10.1039/D0CB00118J
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Very interesting and exciting approach to track antibiotic activity. Thank you for sharing