"Bacterial zorbing": A new mechanism for collective movement of biofilm-like microcolonies
Published in Microbiology
The movement of bacteria either as individual cells or spatiotemporally associated groups, the otherworldly “life at low Reynolds number”1, has continuously fascinated the research community. A better understanding of microscopic life can satisfy human curiosity and may also lead to therapeutic interventions for the pathogens that plague people.
The discovery of this new type of collective movement of bacteria (aka “bacterial zorbing”) was initiated by a conversation between the laboratories of two friends, David J. Beebe and Jo Handelsman, who occupy distinct areas of science: engineering and microbiology. Recently, Chao Li in the Beebe lab developed an under-oil open microfluidic system (UOMS) that enables versatile advanced open-fluid control, open microfluidic devices, and a broad range of applications in experimental biology and biomedicine2,3. Compared to other microfluidic platforms, UOMS provides unprecedented spatiotemporal, optical, and physicochemical access to the cellular or bacterial samples on a device, readily available and easy for biologists to adapt to their own systems. Just as van Leeuwenhoek’s primitive microscope provided the first glimpse of microorganisms, the UOMS tool has offered a new view of bacterial behavior. Soon UOMS found a sweet spot in studying multispecies bacterial communities, one of the research focuses in the Handelsman lab, where Chao Li teamed with Amanda Hurley to discover unexpected corners of microbial life.
Initially, we interrogated a three-membered bacterial community (Pseudomonas aeruginosa, Bacillus cereus, and Flavobacterium johnsoniae4) using UOMS to better visualize the interspecies interactions that modulated biofilm formation of Pseudomonas. On UOMS, we easily have high-quality optical access to the bacterial community microenvironment with single-cell resolution and in situ time-lapse microscopy. From the very first 24-hour time-lapse video, we were surprised by the results. Pseudomonas was not the best biofilm former of the community - Flavobacterium was! In contrast to Flavobacterium’s paltry biofilm formation in 96 well plates, bright, dense multi-cellular puncta emerged from the monolayer of Flavobacterium cells and, to our surprise, the microcolonies were motile. Moreover, the microcolonies grew, moved, and merged with each other over time, forming enormous structures that dispersed simultaneously as if on cue from an invisible conductor. High-magnification microscopy revealed even more surprising details about the mechanism of microcolony movement. Although Neisseria produces microcolonies that merge using pili, the Flavobacterium microcolonies “walk” on a group of cells at the base that orient themselves upright with one pole tethered to the microcolony and the other pole interacting with the surface in a pinwheel movement with repetitive tapping contact on the surface (Figure 1). The motile spherical microcolonies positioned above perpendicular base cells (Movie 1) conjured images of zorbs--inflatable structures into which human thrill-seekers crawl with their legs poking out. And thus, to differentiate them from the pilus-mediated motile microcolonies, we named the Flavobacterium structures “zorbs”.
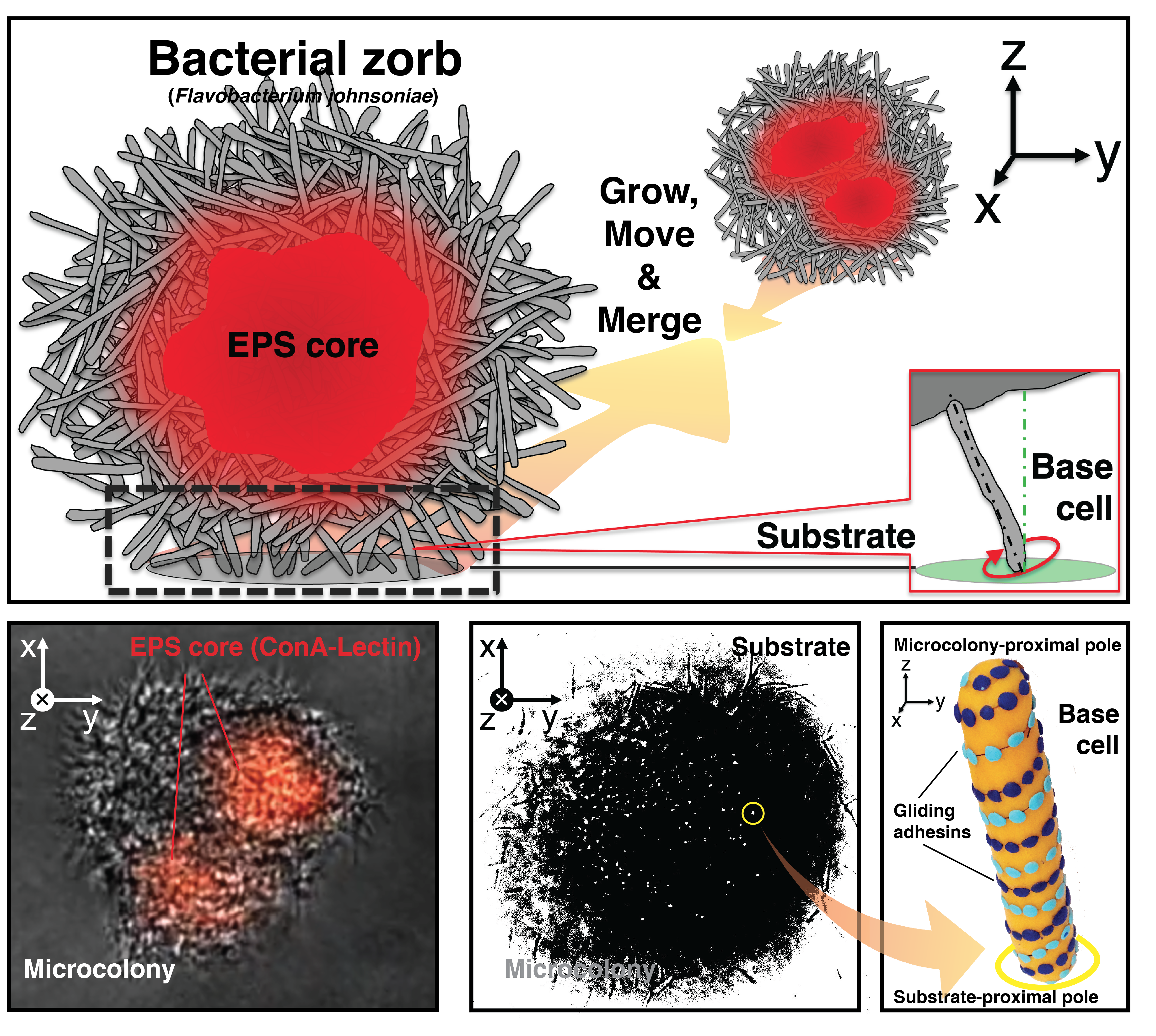
Figure 1. Anatomy and behavior of bacterial zorbs.
We were curious whether the zorb movement was governed by an active process or the result of passive physical forces. To pinpoint the nature of the movement we needed to clarify the energy source. In response to a crucial comment from a reviewer, we applied a proton motive force (PMF) uncoupler to interrupt the bacterial powertrain. As expected for an active mechanism, the PMF inhibitor immediately stopped the movement of the microcolonies as well. Identification of the active movement sparked many interesting questions: Are the microcolonies just a cluster of interacting bacteria or a biofilm-like structure? Could Flavobacterium gliding apparatus contribute to zorb movement? How do the individual bacteria at the base generate and transfer the propulsion force to cause long-range, directional movement?
To peek further into this bizarre bacterial zorbing behavior, we resorted to our collaborators in the Pan mechanical engineering lab that focuses on advanced computational methods, complex fluids, and soft matter. The questions we were trying to answer include how much force may be required to propel a microcolony in such a low Reynolds number fluidic environment, and how might the force generated by a base cell compare with the propulsion force on a microcolony. Thanks to a new computational method5 from the Pan lab based on the generalized moving least squares (GMLS) discretization, we were able to solve the full Stokes equations and thus compute the forces at different levels of the moving objects. While direct measurement of the forces in experiments is still challenging and missing in this work, the simulated results suggested an interesting mechanism by which base cells act together to generate the force to propel microcolonies at the measured velocity.
The discovery of zorbs is a story that illustrates the fruits of multidisciplinary collaborations and following curiosity. This work raises many more questions than it answers, such as What is the biological role of biofilm-like, motile microcolonies? Is there a synchronization (either passive or active) among the base cells to propel the whole structure? How do the bacteria sense and interact with each other to merge zorbs? What signals induce the formation and dispersion of zorbs? How do Flavobacterium zorbs interact with other species in a community? Are zorbs only produced by gliding bacteria? We hope this work intrigues the research community as it does the authors. Finally, we thank our dedicated editors and reviewers that helped us produce a more impactful publication.
- Purcell, E. M. Life at low Reynolds number. AIP Conference Proceedings (1976) doi:10.1063/1.30370.
- Li, C. et al. Double-exclusive liquid repellency (double-ELR): an enabling technology for rare phenotype analysis. Lab Chip 18, 2710–2719 (2018).
- Li, C. et al. Under oil open-channel microfluidics empowered by exclusive liquid repellency. Sci Adv 6, eaay9919 (2020).
- Lozano, G. L. et al. Introducing THOR, a model microbiome for genetic dissection of community behavior. mBio 10, 2 (2019).
- Hu, W., Trask, N., Hu, X. & Pan, W. A spatially adaptive high-order meshless method for fluid–structure interactions. Comput. Methods Appl. Mech. Eng. 355, 67–93 (2019).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Fascinating research and a story well told! Thanks for sharing. I am embarking on the isolation and characterization of surface colonizing bacteria and may be in touch to learn about your fluidics / imaging system.
Dear Roland,
Nice to e-meet you on Nature Portfolio.
Glad to know you find the bacterial zorbing work and our under-oil system interesting. Please just let me know whenever you're available and what to have a chat about bacteria.
Best,
Chao