Epigenetics in Motion: How DNMT1 Steers Interneuron Migration and Identity
Published in Chemistry, Neuroscience, and Physics
Why is the research valuable?
How do neurons know where to go and what they should become? The development of the cerebral cortex depends on the precise migration, positioning, and fate stabilization of its neuronal components: excitatory neurons and inhibitory interneurons. One group of molecules critical in this context are epigenetic modifiers, such as DNA methyltransferases, which regulate gene expression without changing the DNA sequence. Among them, DNMT1 has long been viewed as a maintenance enzyme for DNA methylation during cell division. But what is its role once cells have exited the cell cycle?
We were particularly interested in how DNMT1 functions in postmitotic inhibitory interneurons, specifically, somatostatin-positive (SST+) interneurons that migrate tangentially from the medial ganglionic eminence to the developing cortex. Cortical interneuron dysfunction, often rooted in developmental disturbances, has been linked to a range of neurodevelopmental and neuropsychiatric disorders, including schizophrenia, autism spectrum disorders, and epilepsy. Moreover, DNMT1 dysregulation itself has been implicated in these conditions. Our study provides mechanistic insight into how epigenetic disruption—via DNMT1—can impair interneuron positioning and identity after mitosis, potentially contributing to faulty circuit assembly and disease-relevant behavioral outcomes. More broadly, our work addresses a longstanding question: do epigenetic regulators like DNMT1 actively shape interneuron subtype identity and positioning after cell division, and if so, how might this influence cortical circuit formation and function?
What did the authors do?
This study has been a long journey. It started during the time of my group at the University of Jena, where we first established conditional mouse lines targeting DNMT1 in specific interneuron populations. After relocating to Aachen, we had to rebuild these lines, reacquire licenses, and re-establish our experimental pipelines from scratch. The first striking phenotype we observed was in adult mice: a shift in cortical layering and behavioral abnormalities. But it took much longer—and significant technical effort—to pinpoint the embryonic origin of these changes.
We used an Sst-Cre line to conditionally delete Dnmt1 in SST+ interneurons and combined this with live imaging in embryonic brain slices, transcriptomics, and global DNA methylation profiling. We found that DNMT1-deficient SST+ interneurons prematurely enter the cortical plate, deviating from their stereotypical migratory route along the marginal zone. This altered positioning affects the local environment, including proliferation dynamics of radial glial progenitors, indicating a non-cell-autonomous effect. Here, we received important support from the Tanja Vogel’s group, who provided single cell RNA sequencing data supporting this communication between SST-interneurons and cortical progenitors.
Further transcriptomic analysis revealed the deregulation of key transcription factors such as Arx, which are essential for interneuron identity and migration. Importantly, we observed a partial fate switch toward a parvalbumin-like (PV) expression profile, suggesting that DNMT1 safeguards SST interneuron subtype identity even after cell cycle exit.
To better understand the structural basis of DNMT1’s function in non-dividing cells, molecular modeling and simulation approaches were carried out in collaboration with the team of Paolo Carloni. These revealed that DNMT1 can stably bind to unmethylated DNA, supporting the notion that DNMT1 may have regulatory functions beyond classical maintenance methylation.
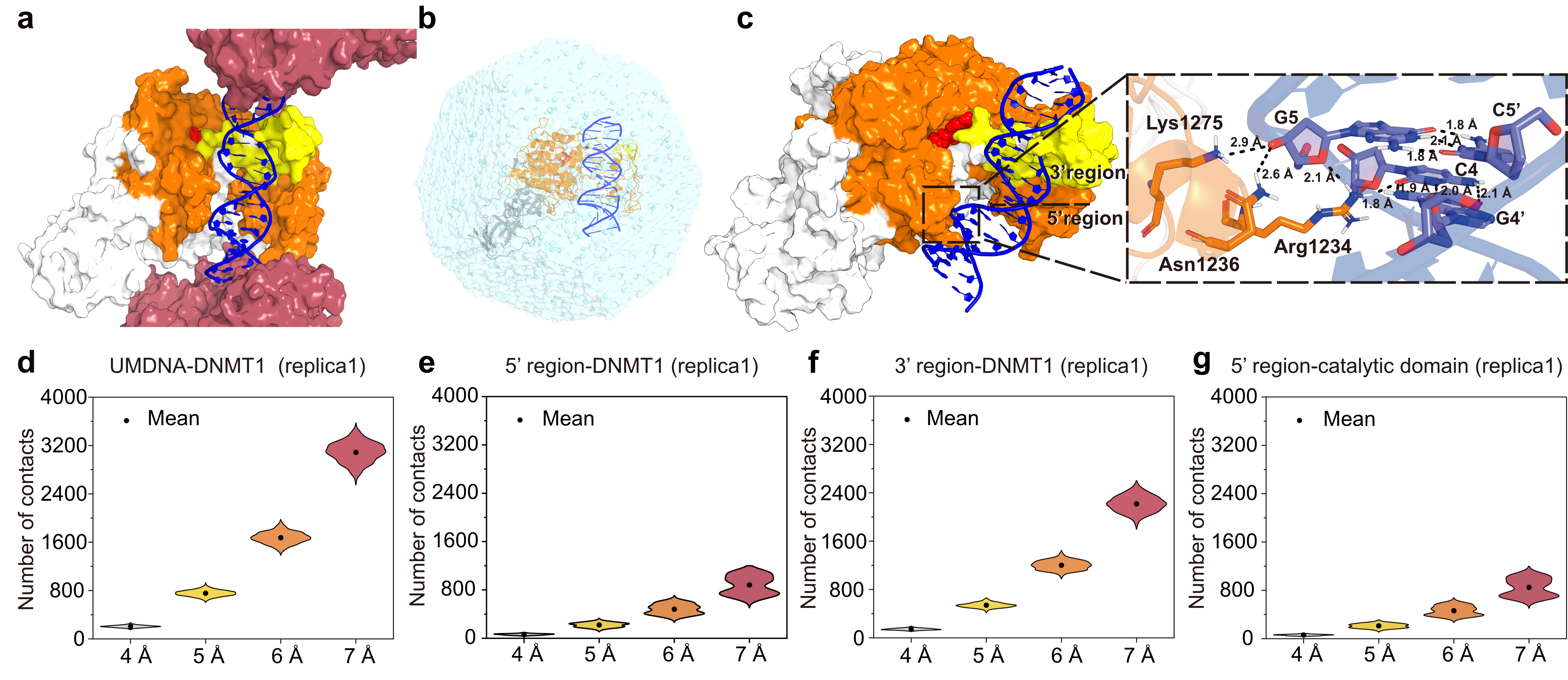
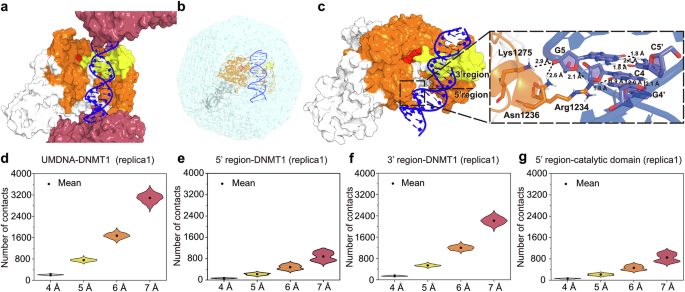
Figure 1 – Molecular dynamic simulations of DNMT1 binding to unmethylated DNA (UMDNA)
(a) DNMT1/UMDNA/SAH complex unit in the X-ray structure (PDB ID: 3PTA). Inter-unit DNMT1 protein regions (rose-pink) are mostly positively charged and interact with the UMDNA (blue). Intra-unit DNMT1 regions include the autoinhibitory linker BAH1/BAH2 (white surface), catalytic domain (orange), SAH (red spheres), and CXXC domain (yellow). (b-g) Simulations of DNMT1/UMDNA interactions in aqueous solution. (b) Illustration of the DNMT1/UMDNA complex in solution. (c) The complex at the end of one of our three MD simulations (same coloring scheme as in (a)), with residues at the CpG site interacting with the catalytic domain highlighted by sticks. Only hydrogen atoms bound to polar groups are shown (as black dashed lines). (d-g)Number of contacts between UMDNA (d), the 5’ (e) and 3’ regions (f) with DNMT1, as well as between the 5’ region and the catalytic domain of DNMT1 (g) during the last-100-ns simulation.
Alongside molecular and imaging work, functional insights were derived from Neuropixels-based in vivo recordings, performed by Simon Musall, which allowed us to relate the observed developmental changes to alterations in cortical activity patterns. The study also benefited from an inspiring exchange of ideas and unpublished data with Tanja Vogel’s group, particularly in the field of epigenetic regulation in neural development.
Just when our experimental infrastructure had finally been re-established, the COVID-19 pandemic hit, delaying animal work and adding logistical complexity. Despite these hurdles, we persevered, and the story slowly came together.
What are the implications of this study?
This study provides new insight into how cortical interneuron development is epigenetically fine-tuned not only at the progenitor level but also postmitotically. It demonstrates that DNMT1 regulates migration and identity acquisition in SST+ interneurons by acting on transcriptional networks even after cell cycle exit, thus extending its known role beyond DNA methylation maintenance. This work supports an emerging model in which interneuron specification is not strictly hard-wired at the progenitor stage but remains plastic and environmentally responsive well into postmitotic life1,2,3. This opens new avenues for exploring how external cues—such as neurotransmitters, hormones, or stress—can shape the epigenetic landscape in a cell-type-specific manner4,5,6.
These findings underscore the importance of spatiotemporally controlled epigenetic regulation during brain development. They also point to novel mechanisms by which interneuron dysfunction—linked to neurodevelopmental and neuropsychiatric disorders such as autism and schizophrenia—may arise. Given the broad relevance of GABAergic circuits in cognition, mood regulation, and oscillatory dynamics, our findings may have implications for understanding the cellular basis of behavioral symptoms in disease.
From a mechanistic point of view, the unexpected ability of DNMT1 to bind unmethylated DNA in postmitotic neurons suggests that it may act as a modulator of transcriptional plasticity, not merely a passive maintenance enzyme. This opens new avenues for investigating DNMT1’s interactions with non-coding RNAs and chromatin remodelers, especially in neuronal subtypes.
Finally, this work highlights the value of interdisciplinary collaboration—from in vivo electrophysiology to molecular modeling—and the power of integrating developmental biology with structural biochemistry and systems neuroscience.
Link to the article:
Reichard, J., Wolff, P., Xie, S. et al. DNMT1-mediated regulation of somatostatin-positive interneuron migration impacts cortical architecture and function. Nat Commun16, 6834 (2025). https://doi.org/10.1038/s41467-025-62114-0
References:
1. Bright, A. R. et al. Temporal control of progenitor competence shapes maturation in GABAergic neuron development in mice. Nat Neurosci (2025) doi:10.1038/s41593-025-01999-y.
2. Mayer, C., Bandler, R. C. & Fishell, G. Lineage Is a Poor Predictor of Interneuron Positioning within the Forebrain. Neuron 92, 45–51 (2016).
3. De Marco García, N. V., Karayannis, T. & Fishell, G. Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature 472, 351–355 (2011).
4. Bagot, R. C., Labonté, B., Peña, C. J. & Nestler, E. J. Epigenetic signaling in psychiatric disorders: stress and depression. Dialogues Clin Neurosci 16, 281–295 (2014).
5. Zhang, T.-Y. & Meaney, M. J. Epigenetics and the Environmental Regulation of the Genome and Its Function. Annu. Rev. Psychol. 61, 439–466 (2010).
6. Guan, J.-S. et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459, 55–60 (2009).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Your space to connect: The Psychedelics Hub
A new Communities’ space to connect, collaborate, and explore research on Psychotherapy, Clinical Psychology, and Neuroscience!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in